Chapter 9 Cross-annotating mouse brains
9.1 Loading the data
We load the classic Zeisel et al. (2015) dataset as our reference. Here, we’ll rely on the fact that the authors have already performed quality control.
We compute log-expression values for use in marker detection inside SingleR().
We examine the distribution of labels in this reference.
##
## (none) Astro1 Astro2 CA1Pyr1 CA1Pyr2 CA1PyrInt CA2Pyr2 Choroid
## 189 68 61 380 447 49 41 10
## ClauPyr Epend Int1 Int10 Int11 Int12 Int13 Int14
## 5 20 12 21 10 21 15 22
## Int15 Int16 Int2 Int3 Int4 Int5 Int6 Int7
## 18 20 24 10 15 20 22 23
## Int8 Int9 Mgl1 Mgl2 Oligo1 Oligo2 Oligo3 Oligo4
## 26 11 17 16 45 98 87 106
## Oligo5 Oligo6 Peric Pvm1 Pvm2 S1PyrDL S1PyrL23 S1PyrL4
## 125 359 21 32 33 81 74 26
## S1PyrL5 S1PyrL5a S1PyrL6 S1PyrL6b SubPyr Vend1 Vend2 Vsmc
## 16 28 39 21 22 32 105 62We load the Tasic et al. (2016) dataset as our test. While not strictly necessary, we remove putative low-quality cells to simplify later interpretation.
sceT <- TasicBrainData()
sceT <- addPerCellQC(sceT, subsets=list(mito=grep("^mt_", rownames(sceT))))
qc <- quickPerCellQC(colData(sceT),
percent_subsets=c("subsets_mito_percent", "altexps_ERCC_percent"))
sceT <- sceT[,which(!qc$discard)]The Tasic dataset was generated using read-based technologies so we need to adjust for the transcript length.
library(AnnotationHub)
mm.db <- AnnotationHub()[["AH73905"]]
mm.exons <- exonsBy(mm.db, by="gene")
mm.exons <- reduce(mm.exons)
mm.len <- sum(width(mm.exons))
mm.symb <- mapIds(mm.db, keys=names(mm.len), keytype="GENEID", column="SYMBOL")
names(mm.len) <- mm.symb
library(scater)
keep <- intersect(names(mm.len), rownames(sceT))
sceT <- sceT[keep,]
assay(sceT, "TPM") <- calculateTPM(sceT, lengths=mm.len[keep])9.2 Applying the annotation
We apply SingleR() with Wilcoxon rank sum test-based marker detection to annotate the Tasic dataset with the Zeisel labels.
library(SingleR)
pred.tasic <- SingleR(test=sceT, ref=sceZ, labels=sceZ$level2class,
assay.type.test="TPM", de.method="wilcox")We examine the distribution of predicted labels:
##
## Astro1 Astro2 CA2Pyr2 Epend Int1 Int10 Int11 Int12
## 1 6 5 1 89 64 2 6
## Int13 Int14 Int15 Int16 Int2 Int3 Int4 Int6
## 9 17 25 8 130 179 30 12
## Int7 Int8 Int9 Oligo1 Oligo2 Oligo3 Oligo4 Oligo6
## 1 77 31 7 1 6 1 1
## Peric S1PyrDL S1PyrL23 S1PyrL4 S1PyrL5a S1PyrL6 S1PyrL6b SubPyr
## 1 354 8 19 200 7 145 4We can also examine the number of discarded cells for each label:
## Lost
## Label FALSE TRUE
## Astro1 1 0
## Astro2 6 0
## CA2Pyr2 4 1
## Epend 1 0
## Int1 89 0
## Int10 64 0
## Int11 2 0
## Int12 5 1
## Int13 9 0
## Int14 17 0
## Int15 25 0
## Int16 8 0
## Int2 129 1
## Int3 176 3
## Int4 29 1
## Int6 12 0
## Int7 1 0
## Int8 77 0
## Int9 31 0
## Oligo1 6 1
## Oligo2 1 0
## Oligo3 6 0
## Oligo4 1 0
## Oligo6 1 0
## Peric 1 0
## S1PyrDL 339 15
## S1PyrL23 8 0
## S1PyrL4 19 0
## S1PyrL5a 200 0
## S1PyrL6 6 1
## S1PyrL6b 145 0
## SubPyr 3 19.3 Diagnostics
We visualize the assignment scores for each label in Figure 9.1.
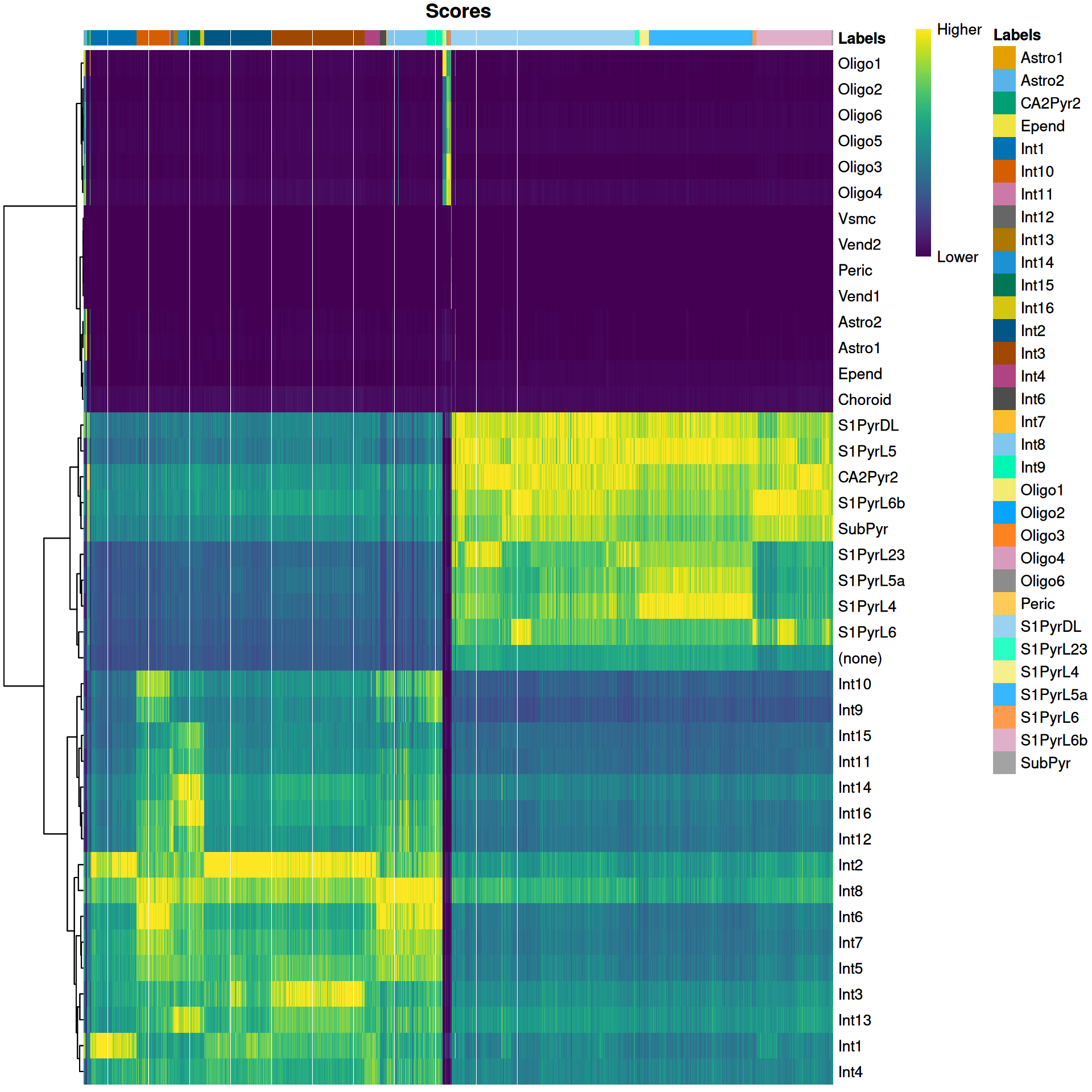
Figure 9.1: Heatmap of the (normalized) assignment scores for each cell (column) in the Tasic test dataset with respect to each label (row) in the Zeisel reference dataset. The final assignment for each cell is shown in the annotation bar at the top.
The delta for each cell is visualized in Figure 9.2.
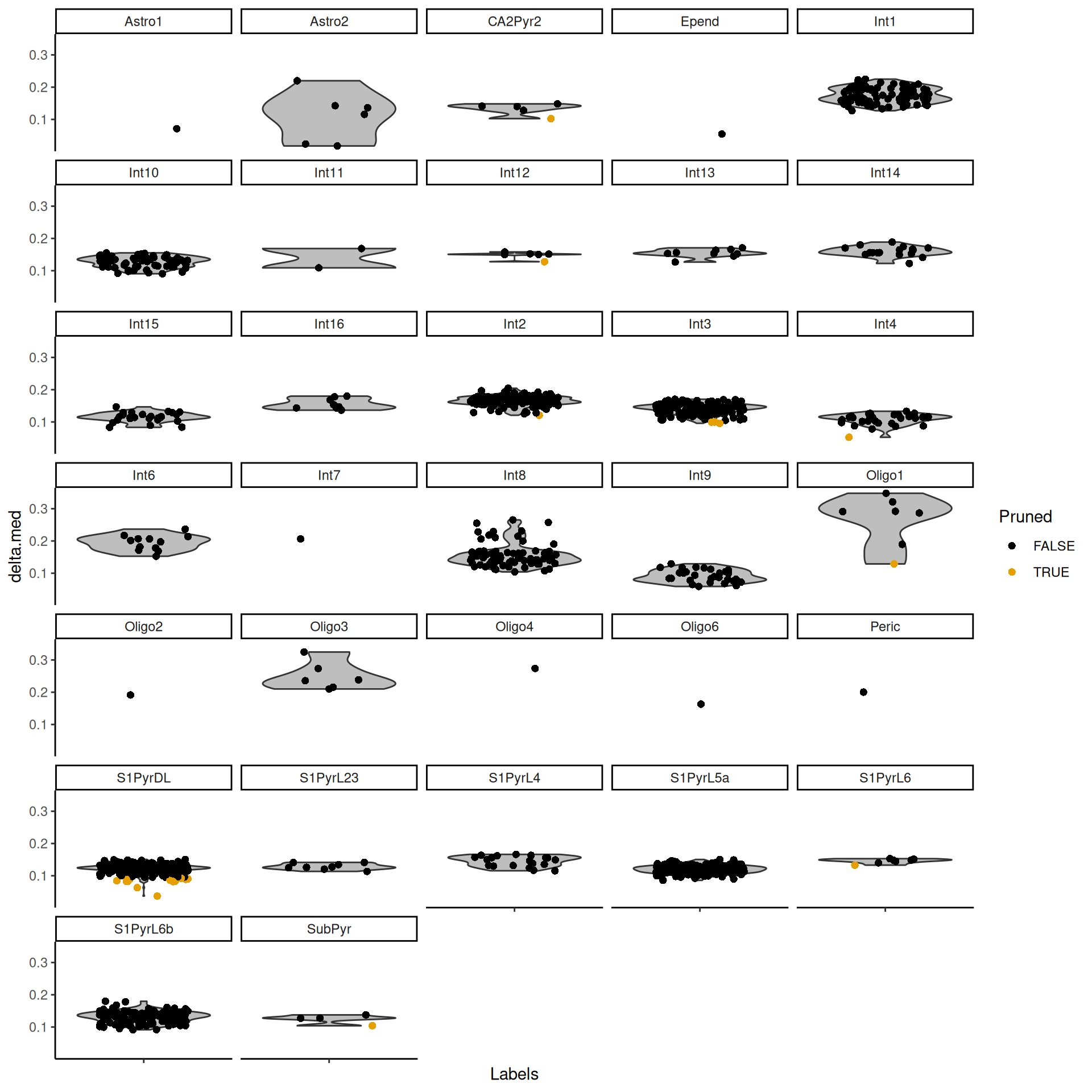
Figure 9.2: Distributions of the deltas for each cell in the Tasic dataset assigned to each label in the Zeisel dataset. Each cell is represented by a point; low-quality assignments that were pruned out are colored in orange.
Finally, we visualize the heatmaps of the marker genes for the most frequent label in Figure 9.3. We could show these for all labels but I wouldn’t want to bore you with a parade of large heatmaps.
library(scater)
collected <- list()
all.markers <- metadata(pred.tasic)$de.genes
sceT <- logNormCounts(sceT)
top.label <- names(sort(table(pred.tasic$labels), decreasing=TRUE))[1]
per.label <- sumCountsAcrossCells(logcounts(sceT),
ids=pred.tasic$labels, average=TRUE)
per.label <- assay(per.label)[unique(unlist(all.markers[[top.label]])),]
pheatmap::pheatmap(per.label, main=top.label)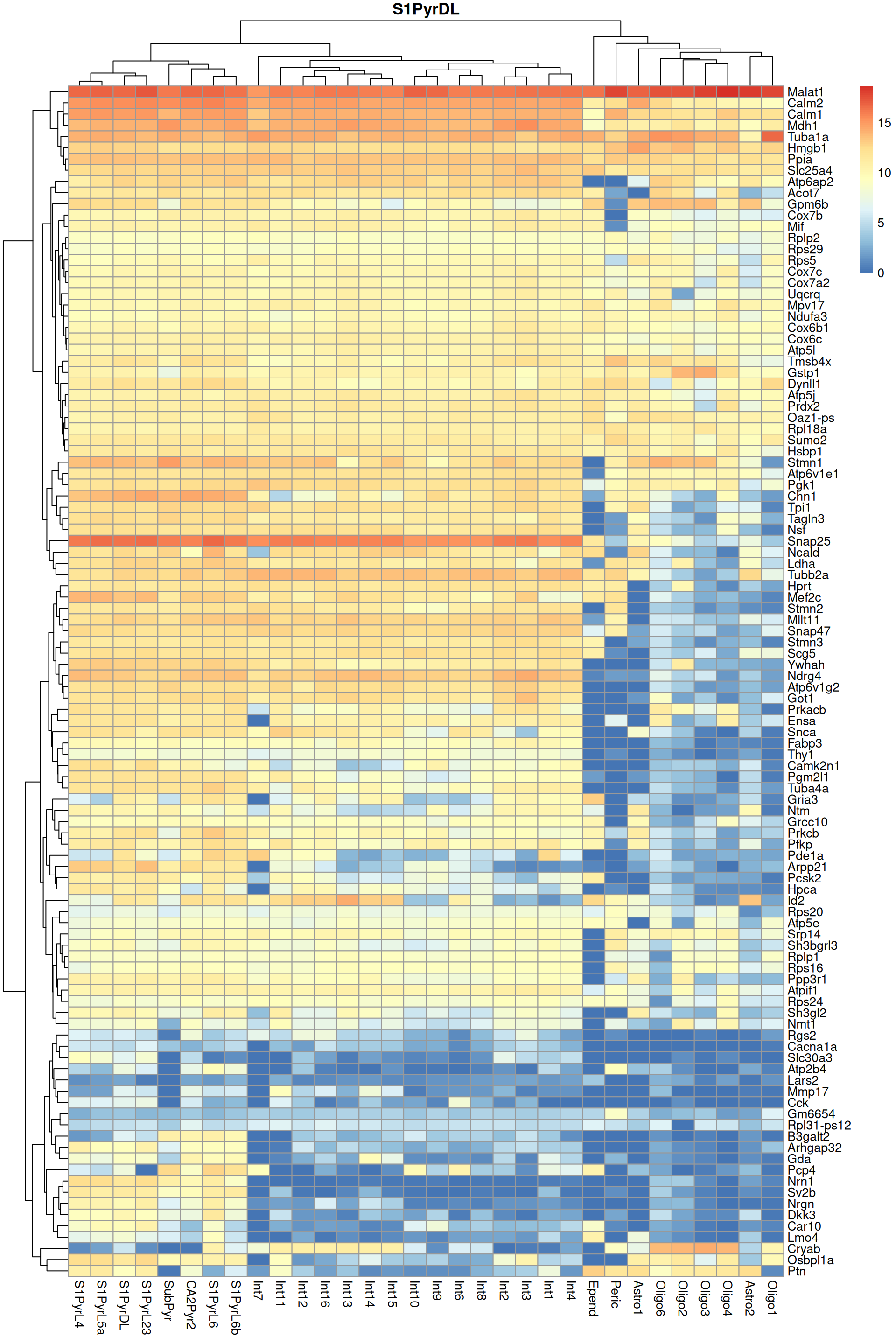
Figure 9.3: Heatmap of log-expression values in the Tasic dataset for all marker genes upregulated in the most frequent label from the Zeisel reference dataset.
9.4 Comparison to clusters
For comparison, we will perform a quick unsupervised analysis of the Grun dataset. We model the variances using the spike-in data and we perform graph-based clustering.
library(scran)
decT <- modelGeneVarWithSpikes(sceT, "ERCC")
set.seed(1000100)
sceT <- denoisePCA(sceT, decT, subset.row=getTopHVGs(decT, n=2500))
library(bluster)
sceT$cluster <- clusterRows(reducedDim(sceT, "PCA"), NNGraphParam())We do not observe a clean 1:1 mapping between clusters and labels in Figure 9.4, probably because many of the labels represent closely related cell types that are difficult to distinguish.
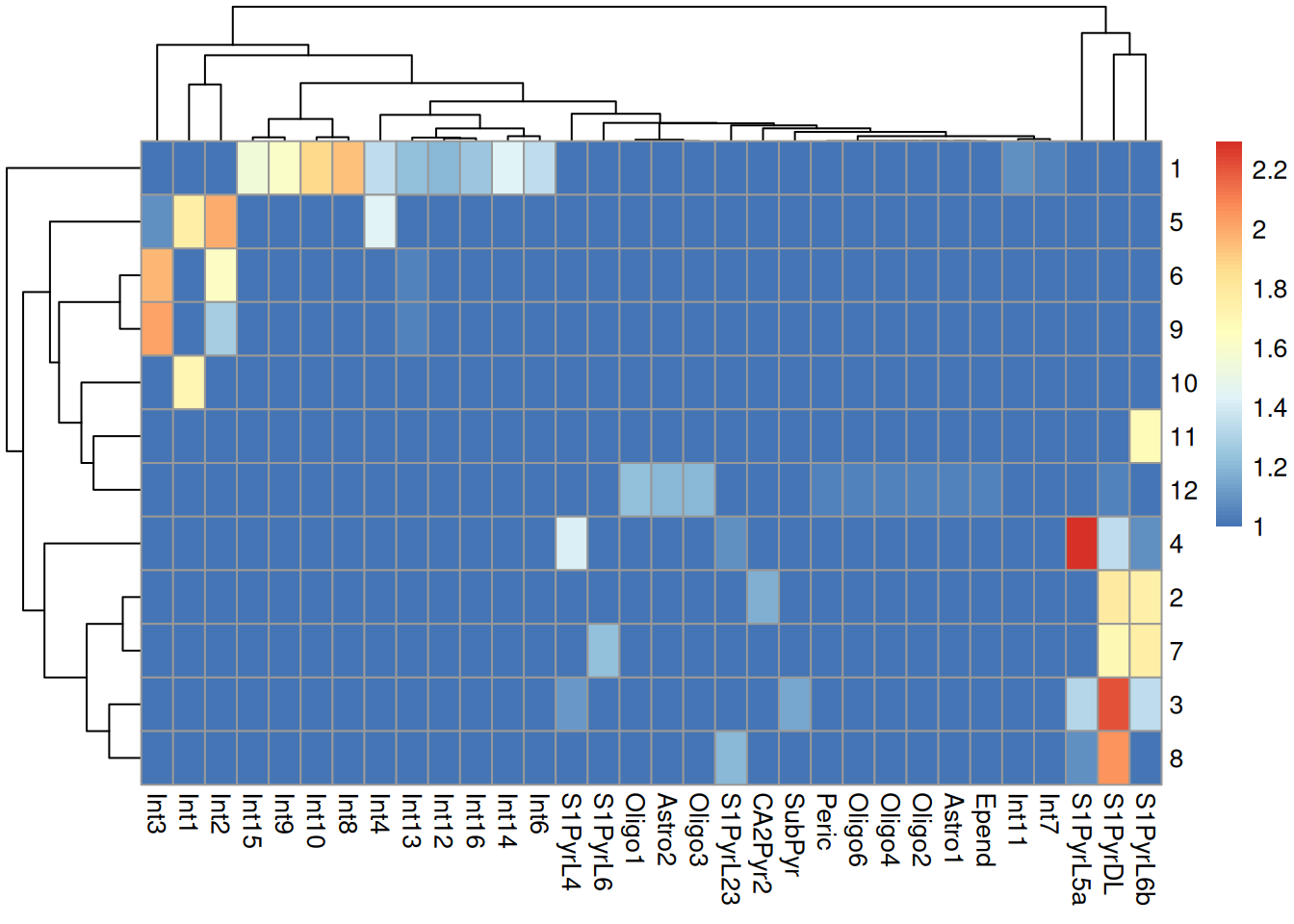
Figure 9.4: Heatmap of the log-transformed number of cells in each combination of label (column) and cluster (row) in the Tasic dataset.
We proceed to the most important part of the analysis. Yes, that’s right, the \(t\)-SNE plot (Figure 9.5).
set.seed(101010100)
sceT <- runTSNE(sceT, dimred="PCA")
plotTSNE(sceT, colour_by="cluster", text_colour="red",
text_by=I(pred.tasic$labels))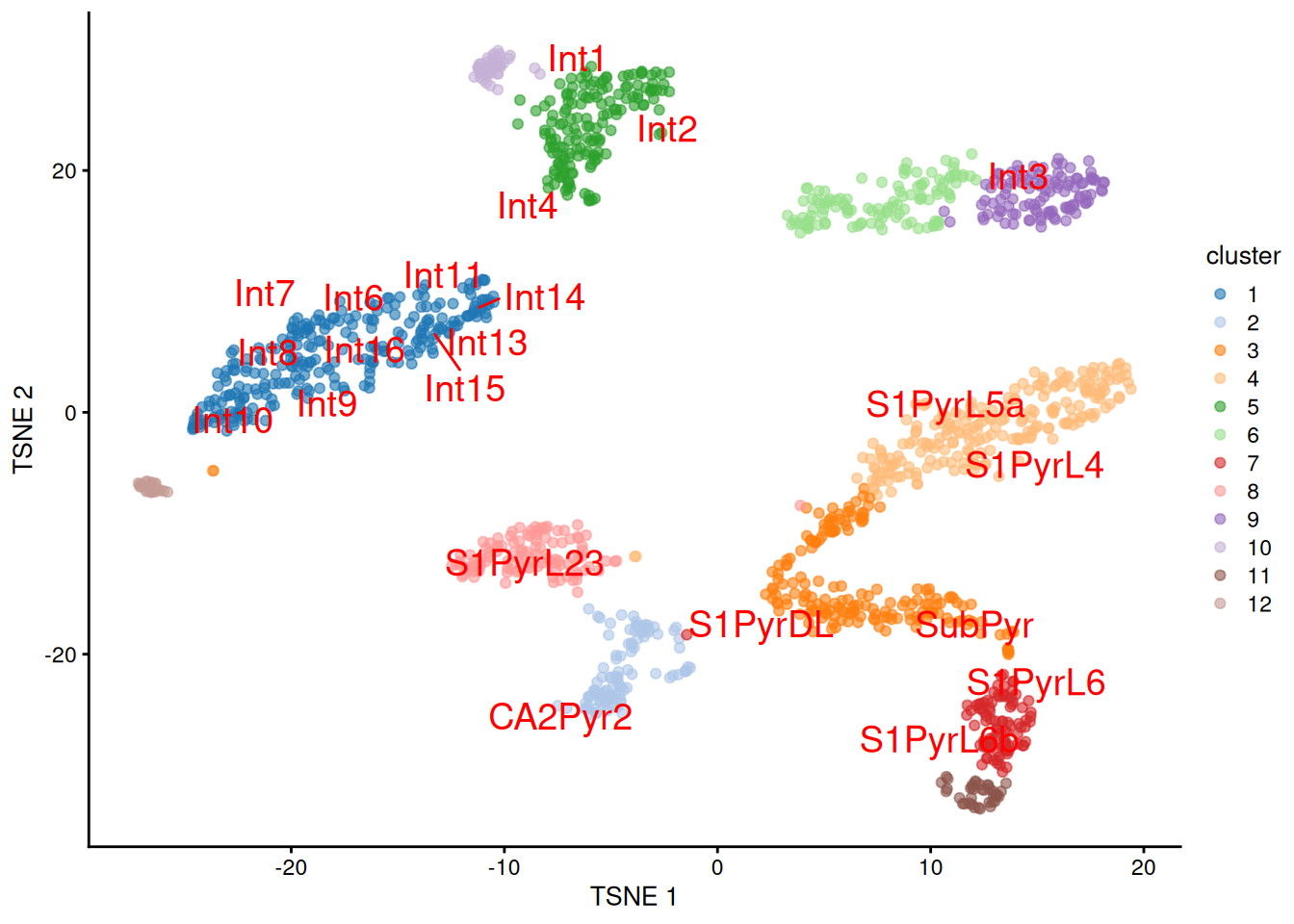
Figure 9.5: \(t\)-SNE plot of the Tasic dataset, where each point is a cell and is colored by the assigned cluster. Reference labels from the Zeisel dataset are also placed on the median coordinate across all cells assigned with that label.
Session information
R Under development (unstable) (2025-10-20 r88955)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 24.04.3 LTS
Matrix products: default
BLAS: /home/biocbuild/bbs-3.23-bioc/R/lib/libRblas.so
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.12.0 LAPACK version 3.12.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_GB LC_COLLATE=C
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: America/New_York
tzcode source: system (glibc)
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] bluster_1.21.0 scran_1.39.0
[3] SingleR_2.13.0 ensembldb_2.35.0
[5] AnnotationFilter_1.35.0 GenomicFeatures_1.63.1
[7] AnnotationDbi_1.73.0 AnnotationHub_4.1.0
[9] BiocFileCache_3.1.0 dbplyr_2.5.1
[11] scater_1.39.0 ggplot2_4.0.0
[13] scuttle_1.21.0 scRNAseq_2.25.0
[15] SingleCellExperiment_1.33.0 SummarizedExperiment_1.41.0
[17] Biobase_2.71.0 GenomicRanges_1.63.0
[19] Seqinfo_1.1.0 IRanges_2.45.0
[21] S4Vectors_0.49.0 BiocGenerics_0.57.0
[23] generics_0.1.4 MatrixGenerics_1.23.0
[25] matrixStats_1.5.0 BiocStyle_2.39.0
[27] rebook_1.21.0
loaded via a namespace (and not attached):
[1] RColorBrewer_1.1-3 jsonlite_2.0.0
[3] CodeDepends_0.6.6 magrittr_2.0.4
[5] ggbeeswarm_0.7.2 gypsum_1.7.0
[7] farver_2.1.2 rmarkdown_2.30
[9] BiocIO_1.21.0 vctrs_0.6.5
[11] DelayedMatrixStats_1.33.0 memoise_2.0.1
[13] Rsamtools_2.27.0 RCurl_1.98-1.17
[15] htmltools_0.5.8.1 S4Arrays_1.11.0
[17] curl_7.0.0 BiocNeighbors_2.5.0
[19] Rhdf5lib_1.33.0 SparseArray_1.11.1
[21] rhdf5_2.55.4 sass_0.4.10
[23] alabaster.base_1.11.1 bslib_0.9.0
[25] alabaster.sce_1.11.0 httr2_1.2.1
[27] cachem_1.1.0 GenomicAlignments_1.47.0
[29] igraph_2.2.1 lifecycle_1.0.4
[31] pkgconfig_2.0.3 rsvd_1.0.5
[33] Matrix_1.7-4 R6_2.6.1
[35] fastmap_1.2.0 digest_0.6.37
[37] dqrng_0.4.1 irlba_2.3.5.1
[39] ExperimentHub_3.1.0 RSQLite_2.4.3
[41] beachmat_2.27.0 labeling_0.4.3
[43] filelock_1.0.3 httr_1.4.7
[45] abind_1.4-8 compiler_4.6.0
[47] bit64_4.6.0-1 withr_3.0.2
[49] S7_0.2.0 BiocParallel_1.45.0
[51] viridis_0.6.5 DBI_1.2.3
[53] HDF5Array_1.39.0 alabaster.ranges_1.11.0
[55] alabaster.schemas_1.11.0 rappdirs_0.3.3
[57] DelayedArray_0.37.0 rjson_0.2.23
[59] tools_4.6.0 vipor_0.4.7
[61] beeswarm_0.4.0 glue_1.8.0
[63] h5mread_1.3.0 restfulr_0.0.16
[65] rhdf5filters_1.23.0 grid_4.6.0
[67] Rtsne_0.17 cluster_2.1.8.1
[69] gtable_0.3.6 metapod_1.19.0
[71] BiocSingular_1.27.0 ScaledMatrix_1.19.0
[73] XVector_0.51.0 ggrepel_0.9.6
[75] BiocVersion_3.23.1 pillar_1.11.1
[77] limma_3.67.0 dplyr_1.1.4
[79] lattice_0.22-7 rtracklayer_1.71.0
[81] bit_4.6.0 tidyselect_1.2.1
[83] locfit_1.5-9.12 Biostrings_2.79.1
[85] knitr_1.50 gridExtra_2.3
[87] scrapper_1.5.1 bookdown_0.45
[89] ProtGenerics_1.43.0 edgeR_4.9.0
[91] xfun_0.54 statmod_1.5.1
[93] pheatmap_1.0.13 UCSC.utils_1.7.0
[95] lazyeval_0.2.2 yaml_2.3.10
[97] evaluate_1.0.5 codetools_0.2-20
[99] cigarillo_1.1.0 tibble_3.3.0
[101] alabaster.matrix_1.11.0 BiocManager_1.30.26
[103] graph_1.89.0 cli_3.6.5
[105] jquerylib_0.1.4 dichromat_2.0-0.1
[107] Rcpp_1.1.0 GenomeInfoDb_1.47.0
[109] dir.expiry_1.19.0 png_0.1-8
[111] XML_3.99-0.19 parallel_4.6.0
[113] blob_1.2.4 sparseMatrixStats_1.23.0
[115] bitops_1.0-9 viridisLite_0.4.2
[117] alabaster.se_1.11.0 scales_1.4.0
[119] purrr_1.2.0 crayon_1.5.3
[121] rlang_1.1.6 cowplot_1.2.0
[123] KEGGREST_1.51.0 Bibliography
Tasic, B., V. Menon, T. N. Nguyen, T. K. Kim, T. Jarsky, Z. Yao, B. Levi, et al. 2016. “Adult mouse cortical cell taxonomy revealed by single cell transcriptomics.” Nat. Neurosci. 19 (2): 335–46.
Zeisel, A., A. B. Munoz-Manchado, S. Codeluppi, P. Lonnerberg, G. La Manno, A. Jureus, S. Marques, et al. 2015. “Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq.” Science 347 (6226): 1138–42.