Chapter 7 Cell type annotation
7.1 Motivation
The most challenging task in scRNA-seq data analysis is arguably the interpretation of the results. Obtaining clusters of cells is fairly straightforward, but it is more difficult to determine what biological state is represented by each of those clusters. Doing so requires us to bridge the gap between the current dataset and prior biological knowledge, and the latter is not always available in a consistent and quantitative manner. Indeed, even the concept of a “cell type” is not clearly defined, with most practitioners possessing a “I’ll know it when I see it” intuition that is not amenable to computational analysis. As such, interpretation of scRNA-seq data is often manual and a common bottleneck in the analysis workflow.
To expedite this step, we can use various computational approaches that exploit prior information to assign meaning to an uncharacterized scRNA-seq dataset. The most obvious sources of prior information are the curated gene sets associated with particular biological processes, e.g., from the Gene Ontology (GO) or the Kyoto Encyclopedia of Genes and Genomes (KEGG) collections. Alternatively, we can directly compare our expression profiles to published reference datasets where each sample or cell has already been annotated with its putative biological state by domain experts. Here, we will demonstrate both approaches with several different scRNA-seq datasets.
7.2 Assigning cell labels from reference data
7.2.1 Overview
A conceptually straightforward annotation approach is to compare the single-cell expression profiles with previously annotated reference datasets. Labels can then be assigned to each cell in our uncharacterized test dataset based on the most similar reference sample(s), for some definition of “similar”. This is a standard classification challenge that can be tackled by standard machine learning techniques such as random forests and support vector machines. Any published and labelled RNA-seq dataset (bulk or single-cell) can be used as a reference, though its reliability depends greatly on the expertise of the original authors who assigned the labels in the first place.
In this section, we will demonstrate the use of the SingleR method (Aran et al. 2019) for cell type annotation. This method assigns labels to cells based on the reference samples with the highest Spearman rank correlations, using only the marker genes between pairs of labels to focus on the relevant differences between cell types. It also performs a fine-tuning step for each cell where the correlations are recomputed with just the marker genes for the top-scoring labels. This aims to resolve any ambiguity between those labels by removing noise from irrelevant markers for other labels. Further details can be found in the SingleR book from which most of the examples here are derived.
7.2.2 Using existing references
For demonstration purposes, we will use one of the 10X PBMC datasets as our test.
While we have already applied quality control, normalization and clustering for this dataset, this is not strictly necessary.
It is entirely possible to run SingleR() on the raw counts without any a priori quality control
and filter on the annotation results at one’s leisure - see the book for an explanation.
#--- loading ---#
library(DropletTestFiles)
raw.path <- getTestFile("tenx-2.1.0-pbmc4k/1.0.0/raw.tar.gz")
out.path <- file.path(tempdir(), "pbmc4k")
untar(raw.path, exdir=out.path)
library(DropletUtils)
fname <- file.path(out.path, "raw_gene_bc_matrices/GRCh38")
sce.pbmc <- read10xCounts(fname, col.names=TRUE)
#--- gene-annotation ---#
library(scater)
rownames(sce.pbmc) <- uniquifyFeatureNames(
rowData(sce.pbmc)$ID, rowData(sce.pbmc)$Symbol)
library(EnsDb.Hsapiens.v86)
location <- mapIds(EnsDb.Hsapiens.v86, keys=rowData(sce.pbmc)$ID,
column="SEQNAME", keytype="GENEID")
#--- cell-detection ---#
set.seed(100)
e.out <- emptyDrops(counts(sce.pbmc))
sce.pbmc <- sce.pbmc[,which(e.out$FDR <= 0.001)]
#--- quality-control ---#
stats <- perCellQCMetrics(sce.pbmc, subsets=list(Mito=which(location=="MT")))
high.mito <- isOutlier(stats$subsets_Mito_percent, type="higher")
sce.pbmc <- sce.pbmc[,!high.mito]
#--- normalization ---#
library(scran)
set.seed(1000)
clusters <- quickCluster(sce.pbmc)
sce.pbmc <- computeSumFactors(sce.pbmc, cluster=clusters)
sce.pbmc <- logNormCounts(sce.pbmc)
#--- variance-modelling ---#
set.seed(1001)
dec.pbmc <- modelGeneVarByPoisson(sce.pbmc)
top.pbmc <- getTopHVGs(dec.pbmc, prop=0.1)
#--- dimensionality-reduction ---#
set.seed(10000)
sce.pbmc <- denoisePCA(sce.pbmc, subset.row=top.pbmc, technical=dec.pbmc)
set.seed(100000)
sce.pbmc <- runTSNE(sce.pbmc, dimred="PCA")
set.seed(1000000)
sce.pbmc <- runUMAP(sce.pbmc, dimred="PCA")
#--- clustering ---#
g <- buildSNNGraph(sce.pbmc, k=10, use.dimred = 'PCA')
clust <- igraph::cluster_walktrap(g)$membership
colLabels(sce.pbmc) <- factor(clust)## class: SingleCellExperiment
## dim: 33694 3985
## metadata(1): Samples
## assays(2): counts logcounts
## rownames(33694): RP11-34P13.3 FAM138A ... AC213203.1 FAM231B
## rowData names(2): ID Symbol
## colnames(3985): AAACCTGAGAAGGCCT-1 AAACCTGAGACAGACC-1 ...
## TTTGTCAGTTAAGACA-1 TTTGTCATCCCAAGAT-1
## colData names(4): Sample Barcode sizeFactor label
## reducedDimNames(3): PCA TSNE UMAP
## mainExpName: NULL
## altExpNames(0):The celldex contains a number of curated reference datasets, mostly assembled from bulk RNA-seq or microarray data of sorted cell types.
These references are often good enough for most applications provided that they contain the cell types that are expected in the test population.
Here, we will use a reference constructed from Blueprint and ENCODE data (Martens and Stunnenberg 2013; The ENCODE Project Consortium 2012);
this is obtained by calling the BlueprintEncode() function to construct a SummarizedExperiment containing log-expression values with curated labels for each sample.
## class: SummarizedExperiment
## dim: 19859 259
## metadata(0):
## assays(1): logcounts
## rownames(19859): TSPAN6 TNMD ... LINC00550 GIMAP1-GIMAP5
## rowData names(0):
## colnames(259): mature.neutrophil
## CD14.positive..CD16.negative.classical.monocyte ...
## epithelial.cell.of.umbilical.artery.1
## dermis.lymphatic.vessel.endothelial.cell.1
## colData names(3): label.main label.fine label.ontWe call the SingleR() function to annotate each of our PBMCs with the main cell type labels from the Blueprint/ENCODE reference.
This returns a DataFrame where each row corresponds to a cell in the test dataset and contains its label assignments.
Alternatively, we could use the labels in ref$label.fine, which provide more resolution at the cost of speed and increased ambiguity in the assignments.
##
## B-cells CD4+ T-cells CD8+ T-cells DC Eosinophils Erythrocytes
## 549 772 1275 1 1 6
## HSC Monocytes NK cells
## 14 1116 251We inspect the results using a heatmap of the per-cell and label scores (Figure 7.1). Ideally, each cell should exhibit a high score in one label relative to all of the others, indicating that the assignment to that label was unambiguous. This is largely the case for monocytes and B cells, whereas we see more ambiguity between CD4+ and CD8+ T cells (and to a lesser extent, NK cells).
![Heatmap of the assignment score for each cell (column) and label (row). Scores are shown before any fine-tuning and are normalized to [0, 1] within each cell.](cell-annotation_files/figure-html/singler-heat-pbmc-1.png)
Figure 7.1: Heatmap of the assignment score for each cell (column) and label (row). Scores are shown before any fine-tuning and are normalized to [0, 1] within each cell.
We compare the assignments with the clustering results to determine the identity of each cluster. Here, several clusters are nested within the monocyte and B cell labels (Figure 7.2), indicating that the clustering represents finer subdivisions within the cell types. Interestingly, our clustering does not effectively distinguish between CD4+ and CD8+ T cell labels. This is probably due to the presence of other factors of heterogeneity within the T cell subpopulation (e.g., activation) that have a stronger influence on unsupervised methods than the a priori expected CD4+/CD8+ distinction.
tab <- table(Assigned=pred$pruned.labels, Cluster=colLabels(sce.pbmc))
# Adding a pseudo-count of 10 to avoid strong color jumps with just 1 cell.
library(pheatmap)
pheatmap(log2(tab+10), color=colorRampPalette(c("white", "blue"))(101))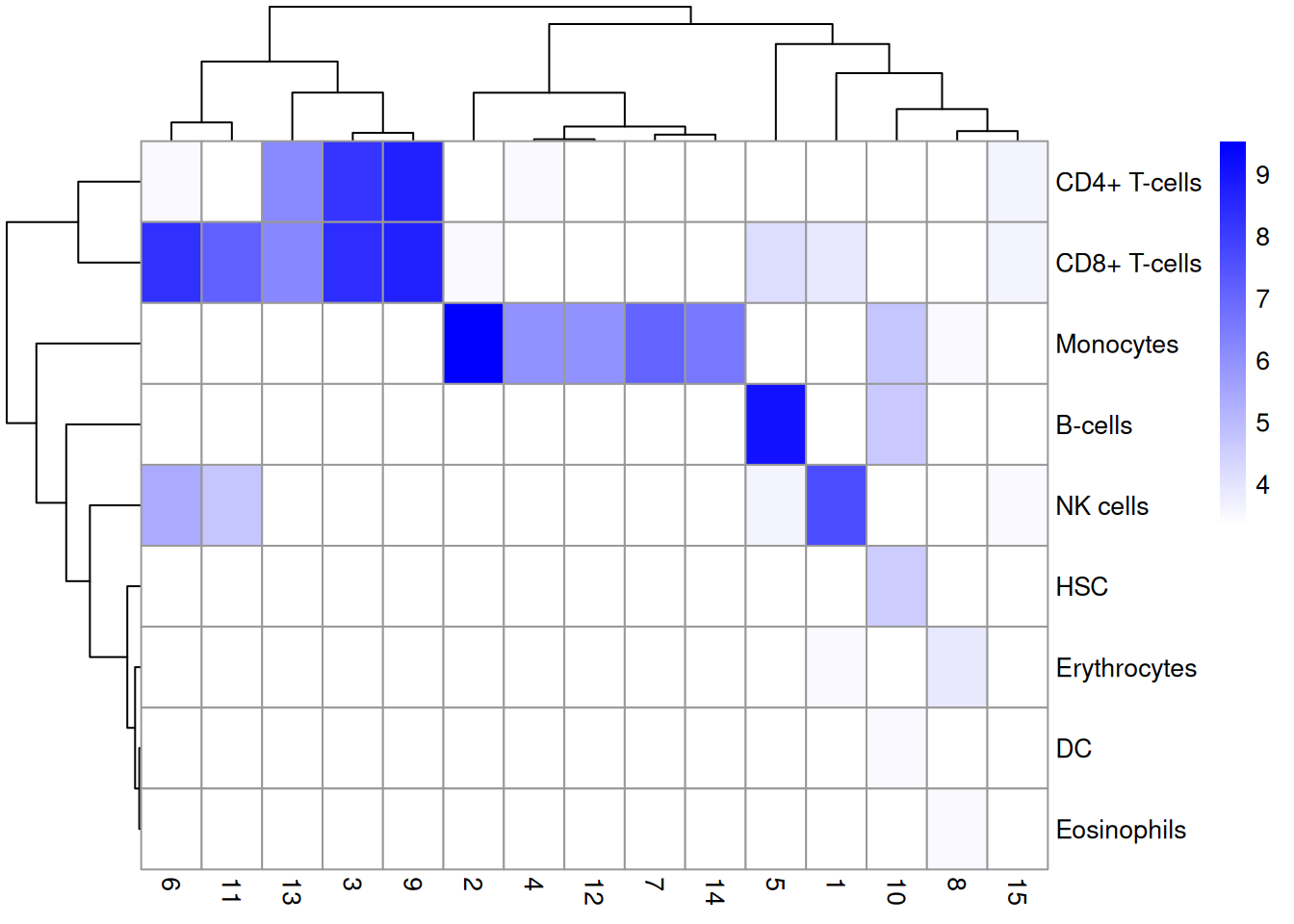
Figure 7.2: Heatmap of the distribution of cells across labels and clusters in the 10X PBMC dataset. Color scale is reported in the log10-number of cells for each cluster-label combination.
This episode highlights some of the differences between reference-based annotation and unsupervised clustering. The former explicitly focuses on aspects of the data that are known to be interesting, simplifying the process of biological interpretation. However, the cost is that the downstream analysis is restricted by the diversity and resolution of the available labels, a problem that is largely avoided by de novo identification of clusters. We suggest applying both strategies to examine the agreement (or lack thereof) between reference label and cluster assignments. Any inconsistencies are not necessarily problematic due to the conceptual differences between the two approaches; indeed, one could use those discrepancies as the basis for further investigation to discover novel factors of variation in the data.
7.2.3 Using custom references
We can also apply SingleR to single-cell reference datasets that are curated and supplied by the user. This is most obviously useful when we have an existing dataset that was previously (manually) annotated and we want to use that knowledge to annotate a new dataset in an automated manner. To illustrate, we will use the Muraro et al. (2016) human pancreas dataset as our reference.
#--- loading ---#
library(scRNAseq)
sce.muraro <- MuraroPancreasData()
#--- gene-annotation ---#
library(AnnotationHub)
edb <- AnnotationHub()[["AH73881"]]
gene.symb <- sub("__chr.*$", "", rownames(sce.muraro))
gene.ids <- mapIds(edb, keys=gene.symb,
keytype="SYMBOL", column="GENEID")
# Removing duplicated genes or genes without Ensembl IDs.
keep <- !is.na(gene.ids) & !duplicated(gene.ids)
sce.muraro <- sce.muraro[keep,]
rownames(sce.muraro) <- gene.ids[keep]
#--- quality-control ---#
library(scater)
stats <- perCellQCMetrics(sce.muraro)
qc <- quickPerCellQC(stats, percent_subsets="altexps_ERCC_percent",
batch=sce.muraro$donor, subset=sce.muraro$donor!="D28")
sce.muraro <- sce.muraro[,!qc$discard]
#--- normalization ---#
library(scran)
set.seed(1000)
clusters <- quickCluster(sce.muraro)
sce.muraro <- computeSumFactors(sce.muraro, clusters=clusters)
sce.muraro <- logNormCounts(sce.muraro)## class: SingleCellExperiment
## dim: 16940 2299
## metadata(0):
## assays(2): counts logcounts
## rownames(16940): ENSG00000268895 ENSG00000121410 ... ENSG00000159840
## ENSG00000074755
## rowData names(2): symbol chr
## colnames(2299): D28-1_1 D28-1_2 ... D30-8_93 D30-8_94
## colData names(4): label donor plate sizeFactor
## reducedDimNames(0):
## mainExpName: endogenous
## altExpNames(1): ERCC# Pruning out unknown or unclear labels.
sce.muraro <- sce.muraro[,!is.na(sce.muraro$label) &
sce.muraro$label!="unclear"]
table(sce.muraro$label)##
## acinar alpha beta delta duct endothelial
## 217 795 442 189 239 18
## epsilon mesenchymal pp
## 3 80 96Our aim is to assign labels to our test dataset from Segerstolpe et al. (2016).
We use the same call to SingleR() but with de.method="wilcox" to identify markers via pairwise Wilcoxon ranked sum tests between labels in the reference Muraro dataset.
This re-uses the same machinery from Chapter 6; further options to fine-tune the test procedure can be passed via the de.args argument.
#--- loading ---#
library(scRNAseq)
sce.seger <- SegerstolpePancreasData()
#--- gene-annotation ---#
library(AnnotationHub)
edb <- AnnotationHub()[["AH73881"]]
symbols <- rowData(sce.seger)$symbol
ens.id <- mapIds(edb, keys=symbols, keytype="SYMBOL", column="GENEID")
ens.id <- ifelse(is.na(ens.id), symbols, ens.id)
# Removing duplicated rows.
keep <- !duplicated(ens.id)
sce.seger <- sce.seger[keep,]
rownames(sce.seger) <- ens.id[keep]
#--- sample-annotation ---#
emtab.meta <- colData(sce.seger)[,c("cell type", "disease",
"individual", "single cell well quality")]
colnames(emtab.meta) <- c("CellType", "Disease", "Donor", "Quality")
colData(sce.seger) <- emtab.meta
sce.seger$CellType <- gsub(" cell", "", sce.seger$CellType)
sce.seger$CellType <- paste0(
toupper(substr(sce.seger$CellType, 1, 1)),
substring(sce.seger$CellType, 2))
#--- quality-control ---#
low.qual <- sce.seger$Quality == "OK, filtered"
library(scater)
stats <- perCellQCMetrics(sce.seger)
qc <- quickPerCellQC(stats, percent_subsets="altexps_ERCC_percent",
batch=sce.seger$Donor,
subset=!sce.seger$Donor %in% c("H6", "H5"))
sce.seger <- sce.seger[,!(qc$discard | low.qual)]
#--- normalization ---#
library(scran)
clusters <- quickCluster(sce.seger)
sce.seger <- computeSumFactors(sce.seger, clusters=clusters)
sce.seger <- logNormCounts(sce.seger) # Converting to FPKM for a more like-for-like comparison to UMI counts.
# However, results are often still good even when this step is skipped.
library(AnnotationHub)
hs.db <- AnnotationHub()[["AH73881"]]
hs.exons <- exonsBy(hs.db, by="gene")
hs.exons <- reduce(hs.exons)
hs.len <- sum(width(hs.exons))
library(scuttle)
available <- intersect(rownames(sce.seger), names(hs.len))
fpkm.seger <- calculateFPKM(sce.seger[available,], hs.len[available])
pred.seger <- SingleR(test=fpkm.seger, ref=sce.muraro,
labels=sce.muraro$label, de.method="wilcox")
table(pred.seger$labels)##
## acinar alpha beta delta duct endothelial
## 191 893 272 106 382 18
## epsilon mesenchymal pp
## 6 52 170As it so happens, we are in the fortunate position where our test dataset also contains independently defined labels. We see strong consistency between the two sets of labels (Figure 7.3), indicating that our automatic annotation is comparable to that generated manually by domain experts.
tab <- table(pred.seger$pruned.labels, sce.seger$CellType)
library(pheatmap)
pheatmap(log2(tab+10), color=colorRampPalette(c("white", "blue"))(101))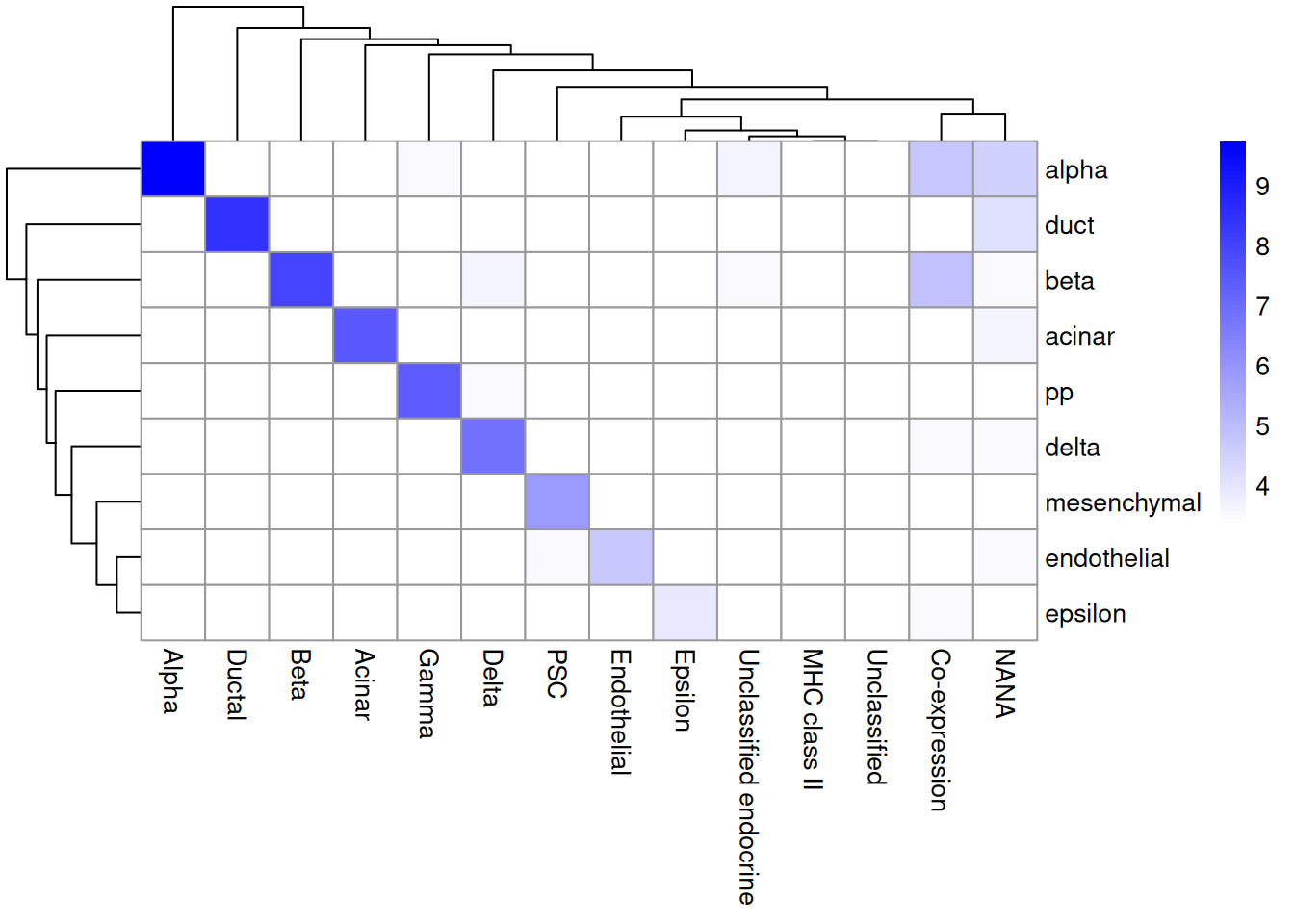
Figure 7.3: Heatmap of the confusion matrix between the predicted labels (rows) and the independently defined labels (columns) in the Segerstolpe dataset. The color is proportinal to the log-transformed number of cells with a given combination of labels from each set.
An interesting question is - given a single-cell reference dataset, is it better to use it directly or convert it to pseudo-bulk values? A single-cell reference preserves the “shape” of the subpopulation in high-dimensional expression space, potentially yielding more accurate predictions when the differences between labels are subtle (or at least capturing ambiguity more accurately to avoid grossly incorrect predictions). However, it also requires more computational work to assign each cell in the test dataset. We refer to the other book for more details on how to achieve a compromise between these two concerns.
7.3 Assigning cell labels from gene sets
A related strategy is to explicitly identify sets of marker genes that are highly expressed in each individual cell. This does not require matching of individual cells to the expression values of the reference dataset, which is faster and more convenient when only the identities of the markers are available. We demonstrate this approach using neuronal cell type markers derived from the Zeisel et al. (2015) study.
#--- loading ---#
library(scRNAseq)
sce.zeisel <- ZeiselBrainData()
library(scater)
sce.zeisel <- aggregateAcrossFeatures(sce.zeisel,
id=sub("_loc[0-9]+$", "", rownames(sce.zeisel)))
#--- gene-annotation ---#
library(org.Mm.eg.db)
rowData(sce.zeisel)$Ensembl <- mapIds(org.Mm.eg.db,
keys=rownames(sce.zeisel), keytype="SYMBOL", column="ENSEMBL")
#--- quality-control ---#
stats <- perCellQCMetrics(sce.zeisel, subsets=list(
Mt=rowData(sce.zeisel)$featureType=="mito"))
qc <- quickPerCellQC(stats, percent_subsets=c("altexps_ERCC_percent",
"subsets_Mt_percent"))
sce.zeisel <- sce.zeisel[,!qc$discard]
#--- normalization ---#
library(scran)
set.seed(1000)
clusters <- quickCluster(sce.zeisel)
sce.zeisel <- computeSumFactors(sce.zeisel, cluster=clusters)
sce.zeisel <- logNormCounts(sce.zeisel)library(scran)
wilcox.z <- pairwiseWilcox(sce.zeisel, sce.zeisel$level1class,
lfc=1, direction="up")
markers.z <- getTopMarkers(wilcox.z$statistics, wilcox.z$pairs,
pairwise=FALSE, n=50)
lengths(markers.z)## astrocytes_ependymal endothelial-mural interneurons
## 79 83 118
## microglia oligodendrocytes pyramidal CA1
## 69 81 125
## pyramidal SS
## 149Our test dataset will be another brain scRNA-seq experiment from Tasic et al. (2016).
## class: SingleCellExperiment
## dim: 24058 1809
## metadata(0):
## assays(1): counts
## rownames(24058): 0610005C13Rik 0610007C21Rik ... mt_X57780 tdTomato
## rowData names(0):
## colnames(1809): Calb2_tdTpositive_cell_1 Calb2_tdTpositive_cell_2 ...
## Rbp4_CTX_250ng_2 Trib2_CTX_250ng_1
## colData names(12): mouse_line cre_driver_1 ... secondary_type
## aibs_vignette_id
## reducedDimNames(0):
## mainExpName: NULL
## altExpNames(1): ERCCWe use the AUCell package to identify marker sets that are highly expressed in each cell. This method ranks genes by their expression values within each cell and constructs a response curve of the number of genes from each marker set that are present with increasing rank. It then computes the area under the curve (AUC) for each marker set, quantifying the enrichment of those markers among the most highly expressed genes in that cell. This is roughly similar to performing a Wilcoxon rank sum test between genes in and outside of the set, but involving only the top ranking genes by expression in each cell.
library(GSEABase)
all.sets <- lapply(names(markers.z), function(x) {
GeneSet(markers.z[[x]], setName=x)
})
all.sets <- GeneSetCollection(all.sets)
library(AUCell)
rankings <- AUCell_buildRankings(counts(sce.tasic),
plotStats=FALSE, verbose=FALSE)
cell.aucs <- AUCell_calcAUC(all.sets, rankings)
results <- t(assay(cell.aucs))
head(results)## gene sets
## cells astrocytes_ependymal endothelial-mural interneurons
## Calb2_tdTpositive_cell_1 0.1387 0.04264 0.5306
## Calb2_tdTpositive_cell_2 0.1366 0.04885 0.4538
## Calb2_tdTpositive_cell_3 0.1087 0.07269 0.3459
## Calb2_tdTpositive_cell_4 0.1322 0.04993 0.5113
## Calb2_tdTpositive_cell_5 0.1513 0.07161 0.4930
## Calb2_tdTpositive_cell_6 0.1342 0.09161 0.3378
## gene sets
## cells microglia oligodendrocytes pyramidal CA1
## Calb2_tdTpositive_cell_1 0.04845 0.1318 0.2318
## Calb2_tdTpositive_cell_2 0.02683 0.1211 0.2063
## Calb2_tdTpositive_cell_3 0.03582 0.1567 0.3219
## Calb2_tdTpositive_cell_4 0.05388 0.1481 0.2547
## Calb2_tdTpositive_cell_5 0.06656 0.1386 0.2088
## Calb2_tdTpositive_cell_6 0.03201 0.1553 0.4011
## gene sets
## cells pyramidal SS
## Calb2_tdTpositive_cell_1 0.3477
## Calb2_tdTpositive_cell_2 0.2762
## Calb2_tdTpositive_cell_3 0.5244
## Calb2_tdTpositive_cell_4 0.3506
## Calb2_tdTpositive_cell_5 0.3010
## Calb2_tdTpositive_cell_6 0.5393We assign cell type identity to each cell in the test dataset by taking the marker set with the top AUC as the label for that cell. Our new labels mostly agree with the original annotation from Tasic et al. (2016), which is encouraging. The only exception involves misassignment of oligodendrocyte precursors to astrocytes, which may be understandable given that they are derived from a common lineage. In the absence of prior annotation, a more general diagnostic check is to compare the assigned labels to cluster identities, under the expectation that most cells of a single cluster would have the same label (or, if multiple labels are present, they should at least represent closely related cell states).
new.labels <- colnames(results)[max.col(results)]
tab <- table(new.labels, sce.tasic$broad_type)
tab##
## new.labels Astrocyte Endothelial Cell GABA-ergic Neuron
## astrocytes_ependymal 43 2 0
## endothelial-mural 0 27 0
## interneurons 0 0 759
## microglia 0 0 0
## oligodendrocytes 0 0 1
## pyramidal SS 0 0 1
##
## new.labels Glutamatergic Neuron Microglia Oligodendrocyte
## astrocytes_ependymal 0 0 0
## endothelial-mural 0 0 0
## interneurons 2 0 0
## microglia 0 22 0
## oligodendrocytes 0 0 38
## pyramidal SS 810 0 0
##
## new.labels Oligodendrocyte Precursor Cell Unclassified
## astrocytes_ependymal 20 4
## endothelial-mural 0 2
## interneurons 0 15
## microglia 0 1
## oligodendrocytes 2 0
## pyramidal SS 0 60As a diagnostic measure, we examine the distribution of AUCs across cells for each label (Figure 7.4). In heterogeneous populations, the distribution for each label should be bimodal with one high-scoring peak containing cells of that cell type and a low-scoring peak containing cells of other types. The gap between these two peaks can be used to derive a threshold for whether a label is “active” for a particular cell. (In this case, we simply take the single highest-scoring label per cell as the labels should be mutually exclusive.) In populations where a particular cell type is expected, lack of clear bimodality for the corresponding label may indicate that its gene set is not sufficiently informative.
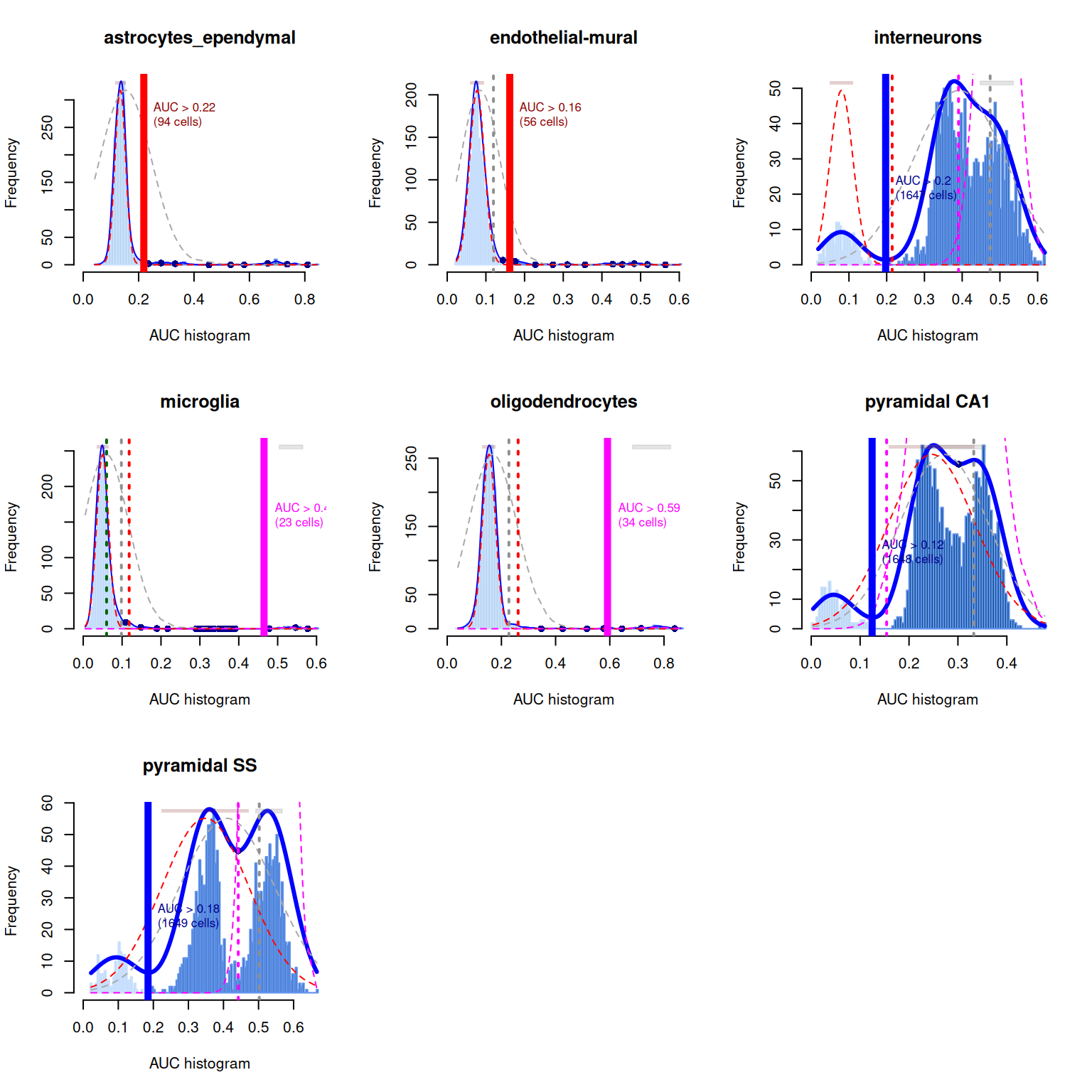
Figure 7.4: Distribution of AUCs in the Tasic brain dataset for each label in the Zeisel dataset. The blue curve represents the density estimate, the red curve represents a fitted two-component mixture of normals, the pink curve represents a fitted three-component mixture, and the grey curve represents a fitted normal distribution. Vertical lines represent threshold estimates corresponding to each estimate of the distribution.
Interpretation of the AUCell results is most straightforward when the marker sets are mutually exclusive, as shown above for the cell type markers. In other applications, one might consider computing AUCs for gene sets associated with signalling or metabolic pathways. It is likely that multiple pathways will be active in any given cell, and it is tempting to use the AUCs to quantify this activity for comparison across cells. However, such comparisons must be interpreted with much caution as the AUCs are competitive values - any increase in one pathway’s activity will naturally reduce the AUCs for all other pathways, potentially resulting in spurious differences across the population.
As we mentioned previously, the advantage of the AUCell approach is that it does not require reference expression values. This is particularly useful when dealing with gene sets derived from the literature or other qualitative forms of biological knowledge. For example, we might instead use single-cell signatures defined from MSigDB, obtained as shown below.
# Downloading the signatures and caching them locally.
library(BiocFileCache)
bfc <- BiocFileCache(ask=FALSE)
scsig.path <- bfcrpath(bfc, file.path("http://software.broadinstitute.org",
"gsea/msigdb/supplemental/scsig.all.v1.0.symbols.gmt"))
scsigs <- getGmt(scsig.path)The flipside is that information on relative expression is lost when only the marker identities are used. The net effect of ignoring expression values is difficult to predict; for example, it may reduce performance for resolving more subtle cell types, but may also improve performance if the per-cell expression was too noisy to be useful. Performance is also highly dependent on the gene sets themselves, which may not be defined in the same context in which they are used. For example, applying all of the MSigDB signatures on the Muraro dataset is rather disappointing (Figure 7.5), while restricting to the subset of pancreas signatures is more promising.
muraro.mat <- counts(sce.muraro)
rownames(muraro.mat) <- rowData(sce.muraro)$symbol
# Explicitly coerce count matrix to a dense matrix to avoid issues with
# support for sparse matrices in AUCell.
muraro.rankings <- AUCell_buildRankings(as.matrix(muraro.mat),
plotStats=FALSE, verbose=FALSE)
# Applying MsigDB to the Muraro dataset, because it's human:
scsig.aucs <- AUCell_calcAUC(scsigs, muraro.rankings)
scsig.results <- t(assay(scsig.aucs))
full.labels <- colnames(scsig.results)[max.col(scsig.results)]
tab <- table(full.labels, sce.muraro$label)
fullheat <- pheatmap(log10(tab+10), color=viridis::viridis(100), silent=TRUE)
# Restricting to the subset of Muraro-derived gene sets:
scsigs.sub <- scsigs[grep("Pancreas", names(scsigs))]
sub.aucs <- AUCell_calcAUC(scsigs.sub, muraro.rankings)
sub.results <- t(assay(sub.aucs))
sub.labels <- colnames(sub.results)[max.col(sub.results)]
tab <- table(sub.labels, sce.muraro$label)
subheat <- pheatmap(log10(tab+10), color=viridis::viridis(100), silent=TRUE)
gridExtra::grid.arrange(fullheat[[4]], subheat[[4]])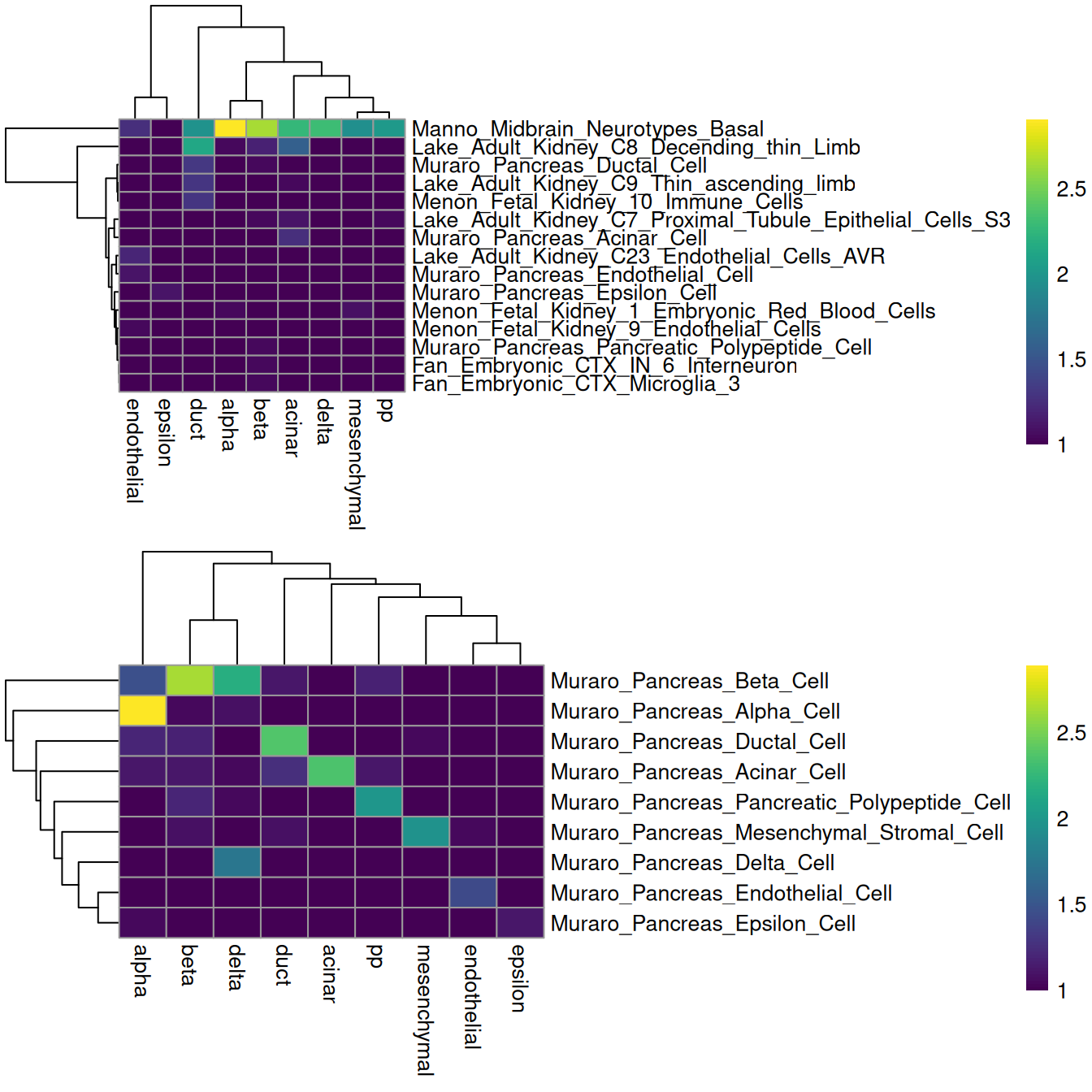
Figure 7.5: Heatmaps of the log-number of cells with each combination of known labels (columns) and assigned MSigDB signatures (rows) in the Muraro data set. The signature assigned to each cell was defined as that with the highest AUC across all (top) or all pancreas-related signatures (bottom).
7.4 Assigning cluster labels from markers
Yet another strategy for annotation is to perform a gene set enrichment analysis on the marker genes defining each cluster. This identifies the pathways and processes that are (relatively) active in each cluster based on upregulation of the associated genes compared to other clusters. We demonstrate on the mouse mammary dataset from Bach et al. (2017), obtaining annotations for the marker genes that define cluster 2. Specifically, we define our marker subset as the top 100 genes with the largest median Cohen’s \(d\) (Chapter 6).
#--- loading ---#
library(scRNAseq)
sce.mam <- BachMammaryData(samples="G_1")
#--- gene-annotation ---#
library(scater)
rownames(sce.mam) <- uniquifyFeatureNames(
rowData(sce.mam)$Ensembl, rowData(sce.mam)$Symbol)
library(AnnotationHub)
ens.mm.v97 <- AnnotationHub()[["AH73905"]]
rowData(sce.mam)$SEQNAME <- mapIds(ens.mm.v97, keys=rowData(sce.mam)$Ensembl,
keytype="GENEID", column="SEQNAME")
#--- quality-control ---#
is.mito <- rowData(sce.mam)$SEQNAME == "MT"
stats <- perCellQCMetrics(sce.mam, subsets=list(Mito=which(is.mito)))
qc <- quickPerCellQC(stats, percent_subsets="subsets_Mito_percent")
sce.mam <- sce.mam[,!qc$discard]
#--- normalization ---#
library(scran)
set.seed(101000110)
clusters <- quickCluster(sce.mam)
sce.mam <- computeSumFactors(sce.mam, clusters=clusters)
sce.mam <- logNormCounts(sce.mam)
#--- variance-modelling ---#
set.seed(00010101)
dec.mam <- modelGeneVarByPoisson(sce.mam)
top.mam <- getTopHVGs(dec.mam, prop=0.1)
#--- dimensionality-reduction ---#
library(BiocSingular)
set.seed(101010011)
sce.mam <- denoisePCA(sce.mam, technical=dec.mam, subset.row=top.mam)
sce.mam <- runTSNE(sce.mam, dimred="PCA")
#--- clustering ---#
snn.gr <- buildSNNGraph(sce.mam, use.dimred="PCA", k=25)
colLabels(sce.mam) <- factor(igraph::cluster_walktrap(snn.gr)$membership)markers.mam <- scoreMarkers(sce.mam, lfc=1)
chosen <- "2"
cur.markers <- markers.mam[[chosen]]
is.de <- order(cur.markers$median.logFC.cohen, decreasing=TRUE)[1:100]
cur.markers[is.de,1:4]## DataFrame with 100 rows and 4 columns
## self.average other.average self.detected other.detected
## <numeric> <numeric> <numeric> <numeric>
## Csn2 8.91711 4.41634 1 0.989444
## Wfdc18 7.85252 3.43375 1 0.835400
## Csn1s1 7.83395 3.71873 1 0.939746
## Muc15 4.80485 1.58288 1 0.447324
## Csn1s2a 9.21574 4.05082 1 0.962808
## ... ... ... ... ...
## Ndrg1 2.16984 1.119877 0.889020 0.538627
## Pycard 1.91299 0.936129 0.893743 0.521984
## Cd14 1.25865 0.711062 0.652893 0.372128
## Ndufa4 3.65888 2.850762 0.995277 0.936963
## Timm13 2.28938 1.561976 0.949233 0.755013We test for enrichment of gene sets defined by the Gene Ontology (GO) project, which describe a comprehensive range of biological processes and functions.
The simplest implementation of this approach involves calling the goana() function from the limma package.
This performs a hypergeometric test to identify GO terms that are overrepresented in our marker subset.
# goana() requires Entrez IDs, some of which map to multiple
# symbols - hence the unique() in the call below.
library(org.Mm.eg.db)
entrez.ids <- mapIds(org.Mm.eg.db, keys=rownames(cur.markers),
column="ENTREZID", keytype="SYMBOL")
library(limma)
go.out <- goana(unique(entrez.ids[is.de]), species="Mm",
universe=unique(entrez.ids))
# Only keeping biological process terms that are not overly general.
go.out <- go.out[order(go.out$P.DE),]
go.useful <- go.out[go.out$Ont=="BP" & go.out$N <= 200,]
head(go.useful[,c(1,3,4)], 30)## Term N DE
## GO:0006641 triglyceride metabolic process 111 7
## GO:1905954 positive regulation of lipid localization 124 7
## GO:0050729 positive regulation of inflammatory response 146 7
## GO:0006639 acylglycerol metabolic process 146 7
## GO:0006638 neutral lipid metabolic process 148 7
## GO:0010884 positive regulation of lipid storage 25 4
## GO:0009060 aerobic respiration 168 7
## GO:0070542 response to fatty acid 71 5
## GO:0015718 monocarboxylic acid transport 186 7
## GO:0032680 regulation of tumor necrosis factor production 187 7
## GO:1905952 regulation of lipid localization 188 7
## GO:1903555 regulation of tumor necrosis factor superfamily cytokine production 190 7
## GO:0032640 tumor necrosis factor production 193 7
## GO:0071706 tumor necrosis factor superfamily cytokine production 196 7
## GO:0019432 triglyceride biosynthetic process 45 4
## GO:0044539 long-chain fatty acid import into cell 19 3
## GO:0019915 lipid storage 108 5
## GO:0055095 lipoprotein particle mediated signaling 4 2
## GO:0055096 low-density lipoprotein particle mediated signaling 4 2
## GO:0071724 response to diacyl bacterial lipopeptide 4 2
## GO:0071726 cellular response to diacyl bacterial lipopeptide 4 2
## GO:0140214 positive regulation of long-chain fatty acid import into cell 4 2
## GO:0046460 neutral lipid biosynthetic process 60 4
## GO:0046463 acylglycerol biosynthetic process 60 4
## GO:0032760 positive regulation of tumor necrosis factor production 114 5
## GO:1903557 positive regulation of tumor necrosis factor superfamily cytokine production 116 5
## GO:0140354 lipid import into cell 24 3
## GO:0006119 oxidative phosphorylation 117 5
## GO:0046470 phosphatidylcholine metabolic process 63 4
## GO:0007263 nitric oxide mediated signal transduction 25 3We see an enrichment for genes involved in lipid storage and lipid synthesis. Given that this is a mammary gland experiment, we might guess that cluster 2 contains luminal epithelial cells responsible for milk production and secretion. Indeed, a closer examination of the marker list indicates that this cluster upregulates milk proteins Csn2 and Csn3 (Figure 7.6).
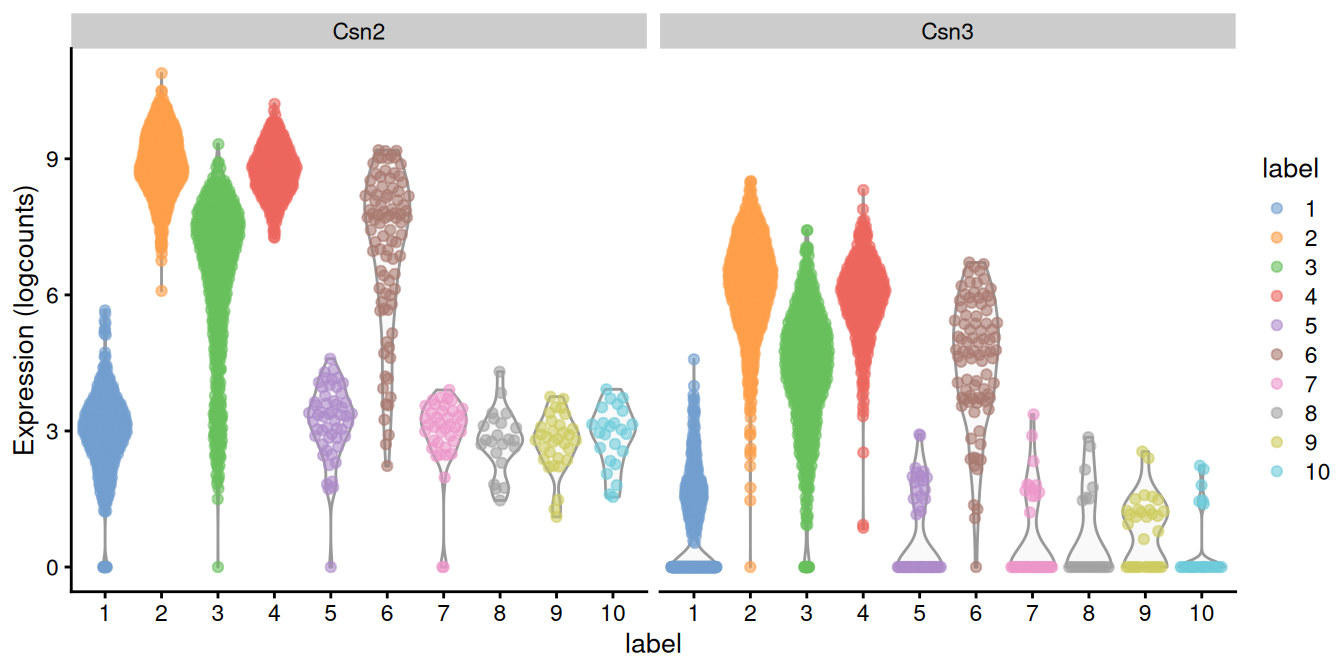
Figure 7.6: Distribution of log-expression values for Csn2 and Csn3 in each cluster.
Further inspection of interesting GO terms is achieved by extracting the relevant genes. This is usually desirable to confirm that the interpretation of the annotated biological process is appropriate. Many terms have overlapping gene sets, so a term may only be highly ranked because it shares genes with a more relevant term that represents the active pathway.
# Extract symbols for each GO term; done once.
tab <- select(org.Mm.eg.db, keytype="SYMBOL", keys=rownames(sce.mam), columns="GOALL")
by.go <- split(tab[,1], tab[,2])
# Identify genes associated with an interesting term.
interesting <- unique(by.go[["GO:0006641"]])
interesting.markers <- cur.markers[rownames(cur.markers) %in% interesting,]
head(interesting.markers[order(-interesting.markers$median.logFC.cohen),1:4], 10)## DataFrame with 10 rows and 4 columns
## self.average other.average self.detected other.detected
## <numeric> <numeric> <numeric> <numeric>
## Dbi 5.85055 3.142174 1.000000 0.911172
## Thrsp 2.72018 0.574604 0.935065 0.270399
## Plb1 2.07492 0.568484 0.860685 0.297144
## Scd1 2.68696 0.920016 0.935065 0.463219
## Acsl1 1.44571 0.419455 0.768595 0.277615
## C3 1.83885 0.706243 0.840614 0.377032
## Lpl 2.11100 1.285142 0.907910 0.525521
## Abhd5 1.05631 0.333045 0.652893 0.250871
## Lipa 1.38386 0.670880 0.763872 0.403527
## Apoc1 1.15912 0.692221 0.646989 0.345021Gene set testing of marker lists is a reliable approach for determining if pathways are up- or down-regulated between clusters. As the top marker genes are simply DEGs, we can directly apply well-established procedures for testing gene enrichment in DEG lists (see here for relevant packages). This contrasts with the AUCell approach where scores are not easily comparable across cells. The downside is that all conclusions are made relative to the other clusters, making it more difficult to determine cell identity if an “outgroup” is not present in the same study.
7.5 Computing gene set activities
For the sake of completeness, we should mention that we can also quantify gene set activity on a per-cell level and test for differences in activity. This inverts the standard gene set testing procedure by combining information across genes first and then testing for differences afterwards. To avoid the pitfalls mentioned previously for the AUCs, we simply compute the average of the log-expression values across all genes in the set for each cell. This is less sensitive to the behavior of other genes in that cell (aside from composition biases, as discussed in Chapter 2).
aggregated <- sumCountsAcrossFeatures(sce.mam, by.go,
exprs_values="logcounts", average=TRUE)
dim(aggregated) # rows are gene sets, columns are cells## [1] 22491 2772## [,1] [,2] [,3] [,4] [,5]
## GO:0000002 0.37093 0.2460 0.1371 0.2772 0.2415
## GO:0000009 0.35356 0.0000 0.0000 0.0000 0.0000
## GO:0000012 0.24200 0.3160 0.1882 0.0000 0.1845
## GO:0000014 0.05303 0.1673 0.2540 0.2663 0.1929
## GO:0000015 0.73220 0.4061 0.4618 0.4842 0.7516
## GO:0000016 0.00000 0.0000 0.0000 0.0000 0.0000
## GO:0000017 0.00000 0.0000 0.0000 0.0000 0.4287
## GO:0000018 0.39482 0.3367 0.1731 0.2765 0.2364
## GO:0000019 0.00000 0.2198 0.0000 0.1614 0.2338
## GO:0000022 1.16894 0.6297 0.0000 1.0441 0.0000We can then identify “differential gene set activity” between clusters by looking for significant differences in the per-set averages of the relevant cells. For example, we observe that cluster 2 has the highest average expression for the triacylglycerol metabolic process GO term (Figure 7.7), consistent with the proposed identity of those cells.
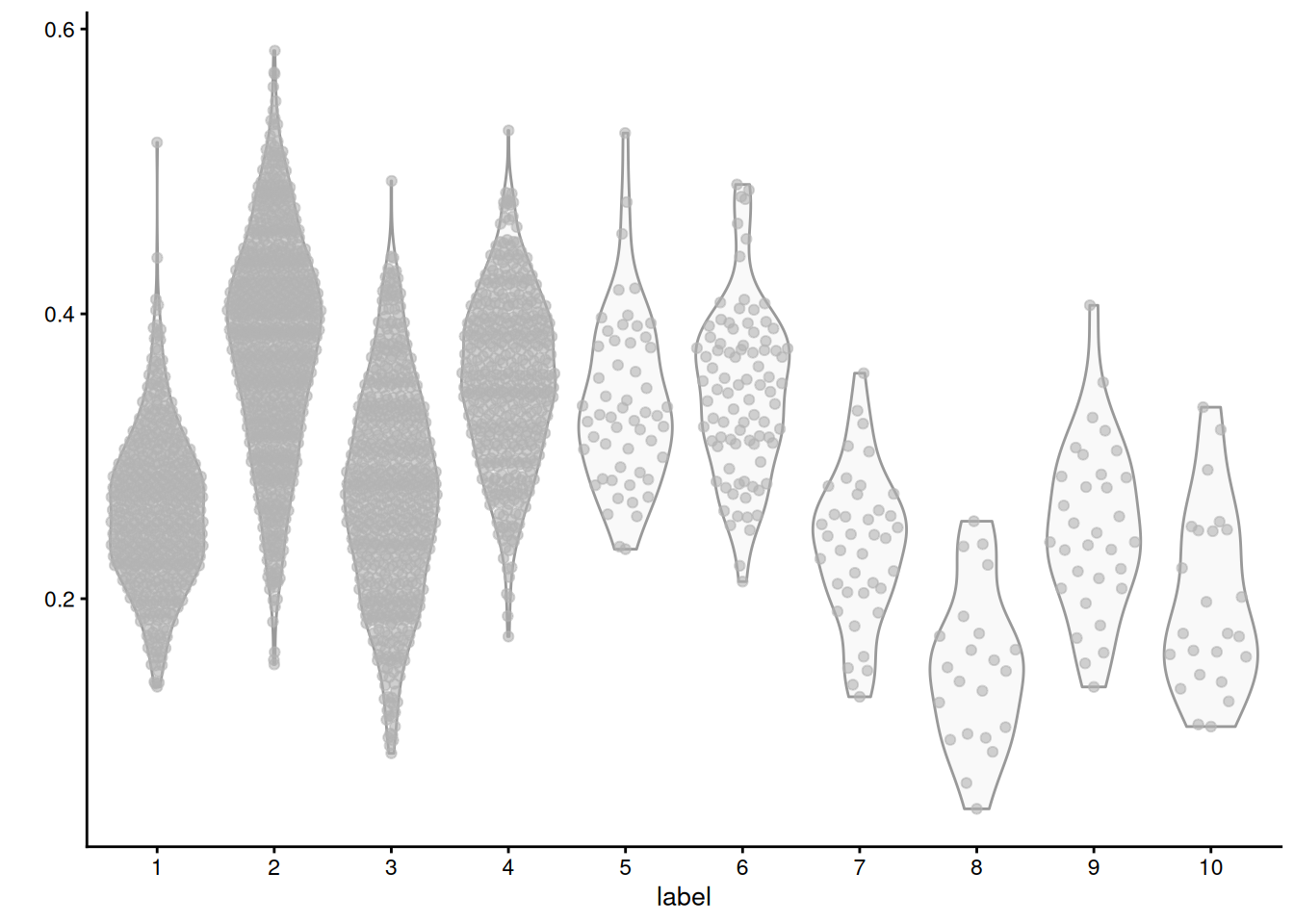
Figure 7.7: Distribution of average log-normalized expression for genes involved in triacylglycerol metabolic process, for all cells in each cluster of the mammary gland dataset.
The obvious disadvantage of this approach is that not all genes in the set may exhibit the same pattern of differences. Non-DE genes will add noise to the per-set average, “diluting” the strength of any differences compared to an analysis that focuses directly on the DE genes (Figure ??). At worst, a gene set may contain subsets of DE genes that change in opposite directions, cancelling out any differences in the per-set average. This is not uncommon for gene sets that contain both positive and negative regulators of a particular biological process or pathway.
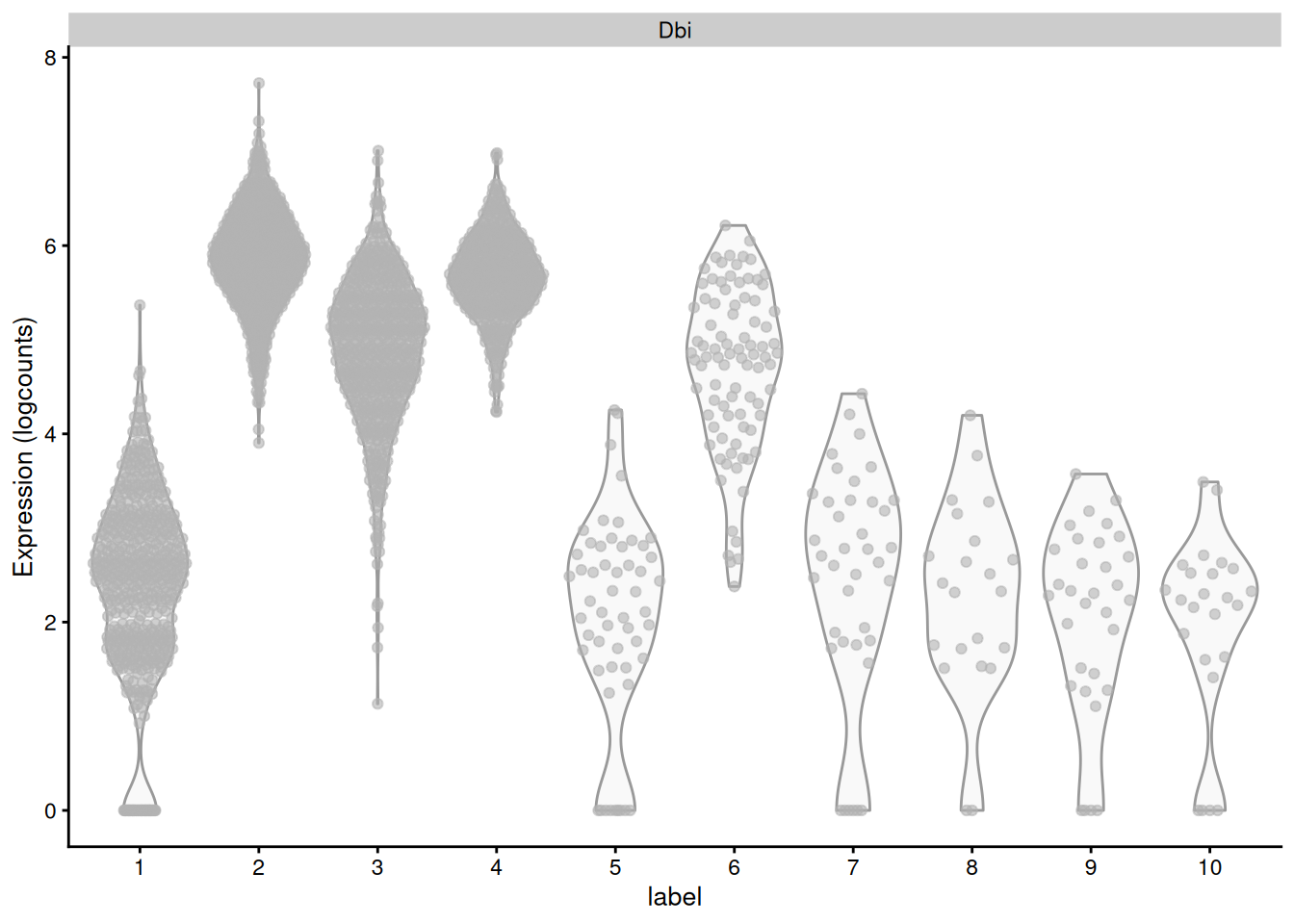
Figure 7.8: Distribution of log-normalized expression values for Dbi across all cells in each cluster of the mammary gland dataset.
We could attempt to use the per-set averages to identify gene sets of interest via differential testing across all possible sets, e.g., with findMarkers().
However, the highest ranking gene sets in this approach tend to be very small and uninteresting because - by definition - the pitfalls mentioned above are avoided when there is only one gene in the set.
This is compounded by the fact that the log-fold changes in the per-set averages are difficult to interpret.
For these reasons, we generally reserve the use of this gene set summary statistic for visualization rather than any real statistical analysis.
Session Info
R version 4.4.2 (2024-10-31)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 24.04.1 LTS
Matrix products: default
BLAS: /home/biocbuild/bbs-3.20-bioc/R/lib/libRblas.so
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.12.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_GB
[4] LC_COLLATE=C LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: America/New_York
tzcode source: system (glibc)
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods base
other attached packages:
[1] scater_1.34.0 ggplot2_3.5.1 limma_3.62.1
[4] org.Mm.eg.db_3.20.0 AUCell_1.28.0 GSEABase_1.68.0
[7] graph_1.84.0 annotate_1.84.0 XML_3.99-0.17
[10] scRNAseq_2.20.0 scran_1.34.0 bluster_1.16.0
[13] scuttle_1.16.0 ensembldb_2.30.0 AnnotationFilter_1.30.0
[16] GenomicFeatures_1.58.0 AnnotationDbi_1.68.0 AnnotationHub_3.14.0
[19] BiocFileCache_2.14.0 dbplyr_2.5.0 pheatmap_1.0.12
[22] SingleR_2.8.0 celldex_1.16.0 SingleCellExperiment_1.28.1
[25] SummarizedExperiment_1.36.0 Biobase_2.66.0 GenomicRanges_1.58.0
[28] GenomeInfoDb_1.42.0 IRanges_2.40.0 S4Vectors_0.44.0
[31] BiocGenerics_0.52.0 MatrixGenerics_1.18.0 matrixStats_1.4.1
[34] BiocStyle_2.34.0 rebook_1.16.0
loaded via a namespace (and not attached):
[1] splines_4.4.2 BiocIO_1.16.0 bitops_1.0-9
[4] filelock_1.0.3 tibble_3.2.1 R.oo_1.27.0
[7] CodeDepends_0.6.6 lifecycle_1.0.4 httr2_1.0.6
[10] edgeR_4.4.0 MASS_7.3-61 lattice_0.22-6
[13] alabaster.base_1.6.1 magrittr_2.0.3 plotly_4.10.4
[16] sass_0.4.9 rmarkdown_2.29 jquerylib_0.1.4
[19] yaml_2.3.10 metapod_1.14.0 cowplot_1.1.3
[22] DBI_1.2.3 RColorBrewer_1.1-3 abind_1.4-8
[25] zlibbioc_1.52.0 purrr_1.0.2 mixtools_2.0.0
[28] R.utils_2.12.3 RCurl_1.98-1.16 rappdirs_0.3.3
[31] GenomeInfoDbData_1.2.13 ggrepel_0.9.6 irlba_2.3.5.1
[34] alabaster.sce_1.6.0 dqrng_0.4.1 DelayedMatrixStats_1.28.0
[37] codetools_0.2-20 DelayedArray_0.32.0 tidyselect_1.2.1
[40] UCSC.utils_1.2.0 farver_2.1.2 ScaledMatrix_1.14.0
[43] viridis_0.6.5 GenomicAlignments_1.42.0 jsonlite_1.8.9
[46] BiocNeighbors_2.0.0 survival_3.7-0 segmented_2.1-3
[49] tools_4.4.2 Rcpp_1.0.13-1 glue_1.8.0
[52] gridExtra_2.3 SparseArray_1.6.0 xfun_0.49
[55] dplyr_1.1.4 HDF5Array_1.34.0 gypsum_1.2.0
[58] withr_3.0.2 BiocManager_1.30.25 fastmap_1.2.0
[61] rhdf5filters_1.18.0 fansi_1.0.6 digest_0.6.37
[64] rsvd_1.0.5 R6_2.5.1 mime_0.12
[67] colorspace_2.1-1 GO.db_3.20.0 RSQLite_2.3.8
[70] R.methodsS3_1.8.2 utf8_1.2.4 tidyr_1.3.1
[73] generics_0.1.3 data.table_1.16.2 rtracklayer_1.66.0
[76] httr_1.4.7 htmlwidgets_1.6.4 S4Arrays_1.6.0
[79] pkgconfig_2.0.3 gtable_0.3.6 blob_1.2.4
[82] XVector_0.46.0 htmltools_0.5.8.1 bookdown_0.41
[85] ProtGenerics_1.38.0 scales_1.3.0 alabaster.matrix_1.6.0
[88] png_0.1-8 knitr_1.49 rjson_0.2.23
[91] nlme_3.1-166 curl_6.0.1 cachem_1.1.0
[94] rhdf5_2.50.0 BiocVersion_3.20.0 vipor_0.4.7
[97] parallel_4.4.2 restfulr_0.0.15 pillar_1.9.0
[100] grid_4.4.2 alabaster.schemas_1.6.0 vctrs_0.6.5
[103] BiocSingular_1.22.0 beachmat_2.22.0 xtable_1.8-4
[106] cluster_2.1.6 beeswarm_0.4.0 evaluate_1.0.1
[109] cli_3.6.3 locfit_1.5-9.10 compiler_4.4.2
[112] Rsamtools_2.22.0 rlang_1.1.4 crayon_1.5.3
[115] labeling_0.4.3 ggbeeswarm_0.7.2 viridisLite_0.4.2
[118] alabaster.se_1.6.0 BiocParallel_1.40.0 munsell_0.5.1
[121] Biostrings_2.74.0 lazyeval_0.2.2 Matrix_1.7-1
[124] dir.expiry_1.14.0 ExperimentHub_2.14.0 sparseMatrixStats_1.18.0
[127] bit64_4.5.2 Rhdf5lib_1.28.0 KEGGREST_1.46.0
[130] statmod_1.5.0 alabaster.ranges_1.6.0 kernlab_0.9-33
[133] igraph_2.1.1 memoise_2.0.1 bslib_0.8.0
[136] bit_4.5.0 Anders, S., and W. Huber. 2010. “Differential expression analysis for sequence count data.” Genome Biol. 11 (10): R106.
Aran, D., A. P. Looney, L. Liu, E. Wu, V. Fong, A. Hsu, S. Chak, et al. 2019. “Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage.” Nat. Immunol. 20 (2): 163–72.
Bach, K., S. Pensa, M. Grzelak, J. Hadfield, D. J. Adams, J. C. Marioni, and W. T. Khaled. 2017. “Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing.” Nat Commun 8 (1): 2128.
Frey, B. J., and D. Dueck. 2007. “Clustering by passing messages between data points.” Science 315 (5814): 972–76.
Glare, E. M., M. Divjak, M. J. Bailey, and E. H. Walters. 2002. “beta-Actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels.” Thorax 57 (9): 765–70.
Guimaraes, J. C., and M. Zavolan. 2016. “Patterns of ribosomal protein expression specify normal and malignant human cells.” Genome Biol. 17 (1): 236.
Ilicic, T., J. K. Kim, A. A. Kołodziejczyk, F. O. Bagger, D. J. McCarthy, J. C. Marioni, and S. A. Teichmann. 2016. “Classification of low quality cells from single-cell RNA-seq data.” Genome Biol. 17 (1): 29.
Islam, S., A. Zeisel, S. Joost, G. La Manno, P. Zajac, M. Kasper, P. Lonnerberg, and S. Linnarsson. 2014. “Quantitative single-cell RNA-seq with unique molecular identifiers.” Nat. Methods 11 (2): 163–66.
Ji, Z., and H. Ji. 2016. “TSCAN: Pseudo-time reconstruction and evaluation in single-cell RNA-seq analysis.” Nucleic Acids Res. 44 (13): e117.
Johnstone, I. M., and A. Y. Lu. 2009. “On Consistency and Sparsity for Principal Components Analysis in High Dimensions.” J Am Stat Assoc 104 (486): 682–93.
Langfelder, P., and S. Horvath. 2007. “Eigengene networks for studying the relationships between co-expression modules.” BMC Syst Biol 1 (November): 54.
Love, M. I., W. Huber, and S. Anders. 2014. “Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2.” Genome Biol. 15 (12): 550.
Lun, A. 2018. “Overcoming Systematic Errors Caused by Log-Transformation of Normalized Single-Cell Rna Sequencing Data.” bioRxiv.
Lun, A. T., K. Bach, and J. C. Marioni. 2016. “Pooling across cells to normalize single-cell RNA sequencing data with many zero counts.” Genome Biol. 17 (April): 75.
Lun, A. T. L., F. J. Calero-Nieto, L. Haim-Vilmovsky, B. Gottgens, and J. C. Marioni. 2017. “Assessing the reliability of spike-in normalization for analyses of single-cell RNA sequencing data.” Genome Res. 27 (11): 1795–1806.
Lun, A. T. L., D. J. McCarthy, and J. C. Marioni. 2016. “A Step-by-Step Workflow for Low-Level Analysis of Single-Cell RNA-seq Data.” F1000Res. 5 (August).
Martens, J. H., and H. G. Stunnenberg. 2013. “BLUEPRINT: mapping human blood cell epigenomes.” Haematologica 98 (10): 1487–9.
McCarthy, D. J., K. R. Campbell, A. T. Lun, and Q. F. Wills. 2017. “Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R.” Bioinformatics 33 (8): 1179–86.
McInnes, Leland, John Healy, and James Melville. 2018. “UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction.” arXiv E-Prints, February, arXiv:1802.03426. http://arxiv.org/abs/1802.03426.
Muraro, M. J., G. Dharmadhikari, D. Grun, N. Groen, T. Dielen, E. Jansen, L. van Gurp, et al. 2016. “A Single-Cell Transcriptome Atlas of the Human Pancreas.” Cell Syst 3 (4): 385–94.
Nazari, F., A. Parham, and A. F. Maleki. 2015. “GAPDH, -actin and -microglobulin, as three common reference genes, are not reliable for gene expression studies in equine adipose- and marrow-derived mesenchymal stem cells.” J Anim Sci Technol 57: 18.
Richard, A. C., A. T. L. Lun, W. W. Y. Lau, B. Gottgens, J. C. Marioni, and G. M. Griffiths. 2018. “T cell cytolytic capacity is independent of initial stimulation strength.” Nat. Immunol. 19 (8): 849–58.
Robinson, M. D., and A. Oshlack. 2010. “A scaling normalization method for differential expression analysis of RNA-seq data.” Genome Biol. 11 (3): R25.
Segerstolpe, A., A. Palasantza, P. Eliasson, E. M. Andersson, A. C. Andreasson, X. Sun, S. Picelli, et al. 2016. “Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes.” Cell Metab. 24 (4): 593–607.
Stegle, O., S. A. Teichmann, and J. C. Marioni. 2015. “Computational and analytical challenges in single-cell transcriptomics.” Nat. Rev. Genet. 16 (3): 133–45.
Tasic, B., V. Menon, T. N. Nguyen, T. K. Kim, T. Jarsky, Z. Yao, B. Levi, et al. 2016. “Adult mouse cortical cell taxonomy revealed by single cell transcriptomics.” Nat. Neurosci. 19 (2): 335–46.
The ENCODE Project Consortium. 2012. “An integrated encyclopedia of DNA elements in the human genome.” Nature 489 (7414): 57–74.
Van der Maaten, L., and G. Hinton. 2008. “Visualizing Data Using T-SNE.” J. Mach. Learn. Res. 9 (2579-2605): 85.
Xu, C., and Z. Su. 2015. “Identification of cell types from single-cell transcriptomes using a novel clustering method.” Bioinformatics 31 (12): 1974–80.
Zeisel, A., A. B. Munoz-Manchado, S. Codeluppi, P. Lonnerberg, G. La Manno, A. Jureus, S. Marques, et al. 2015. “Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq.” Science 347 (6226): 1138–42.
References
Aran, D., A. P. Looney, L. Liu, E. Wu, V. Fong, A. Hsu, S. Chak, et al. 2019. “Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage.” Nat. Immunol. 20 (2): 163–72.
Bach, K., S. Pensa, M. Grzelak, J. Hadfield, D. J. Adams, J. C. Marioni, and W. T. Khaled. 2017. “Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing.” Nat Commun 8 (1): 2128.
Martens, J. H., and H. G. Stunnenberg. 2013. “BLUEPRINT: mapping human blood cell epigenomes.” Haematologica 98 (10): 1487–9.
Muraro, M. J., G. Dharmadhikari, D. Grun, N. Groen, T. Dielen, E. Jansen, L. van Gurp, et al. 2016. “A Single-Cell Transcriptome Atlas of the Human Pancreas.” Cell Syst 3 (4): 385–94.
Segerstolpe, A., A. Palasantza, P. Eliasson, E. M. Andersson, A. C. Andreasson, X. Sun, S. Picelli, et al. 2016. “Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes.” Cell Metab. 24 (4): 593–607.
Tasic, B., V. Menon, T. N. Nguyen, T. K. Kim, T. Jarsky, Z. Yao, B. Levi, et al. 2016. “Adult mouse cortical cell taxonomy revealed by single cell transcriptomics.” Nat. Neurosci. 19 (2): 335–46.
The ENCODE Project Consortium. 2012. “An integrated encyclopedia of DNA elements in the human genome.” Nature 489 (7414): 57–74.
Zeisel, A., A. B. Munoz-Manchado, S. Codeluppi, P. Lonnerberg, G. La Manno, A. Jureus, S. Marques, et al. 2015. “Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq.” Science 347 (6226): 1138–42.