swaglm Overview

swaglm Overview

The swaglm package is a fast implementation of the
Sparse Wrapper Algorithm (SWAG) for Generalized Linear Models (GLM).
SWAG is a meta-learning procedure that combines screening and wrapper
methods to efficiently find strong low-dimensional attribute
combinations for prediction. Additionally, the package provides a
statistical test to assess whether the selected models (learners)
extract meaningful information from the data.
swaglm_test) to determine if the obtained models capture
meaningful structure in the data.Below are instructions on how to install and make use of the
swaglm package.
The swaglm package is currently only available on
GitHub.
# Install dependencies
install.packages(c("devtools"))
# Install/Update the package from GitHub
devtools::install_github("SMAC-Group/swaglm")
# Install the package with Vignettes/User Guides
devtools::install_github("SMAC-Group/swaglm", build_vignettes = TRUE)R librariesThe swaglm package relies on a limited number of
external libraries, but notably on Rcpp and
RcppArmadillo which require a C++ compiler for
installation, such as for example gcc.
library(swaglm)# Simulated data
n <- 2000
p <- 50
X <- MASS::mvrnorm(n = n, mu = rep(0, p), Sigma = diag(rep(1/p, p)))
beta <- c(-15, -10, 5, 10, 15, rep(0, p-5))
# generate from logistic regression model
z <- 1 + X %*% beta
pr <- 1 / (1 + exp(-z))
set.seed(12345)
y <- as.factor(rbinom(n, 1, pr))
y <- as.numeric(y) - 1
# Run SWAG
swaglm_obj <- swaglm(X = X, y = y, p_max = 20, family = binomial(),
alpha = 0.15, verbose = TRUE, seed = 123)## Completed models of dimension 1
## Completed models of dimension 2
## Completed models of dimension 3
## Completed models of dimension 4
## Completed models of dimension 5
## Completed models of dimension 6
## Completed models of dimension 7
## Completed models of dimension 8print(swaglm_obj)## SWAGLM results :
## -----------------------------------------
## Input matrix dimension: 2000 50
## Number of explored models: 136
## Number of dimensions explored: 8# plot network
swaglm_network_obj = compute_network(swaglm_obj)
plot(swaglm_network_obj, scale_vertex = 1)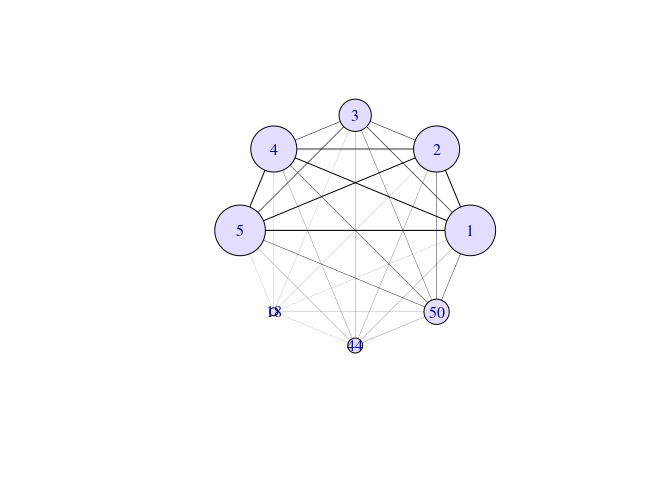
# Run statistical test
B=20
test_results <- swaglm_test(swaglm_obj, B = B, verbose = TRUE)
# View p-values for both entropy-based measures
print(test_results)## SWAGLM Test Results:
## ----------------------
## p-value (Eigen): 0.1306
## p-value (Freq): 0Find vignettes with detailed examples as well as the user’s manual at the package website.
The function swaglm_test() performs a permutation test
to evaluate whether the selected variables contain meaningful
information or are randomly selected.
Null Hypothesis: The selected models are no different from randomly chosen ones.
The response variable is shuffled to break its true relationship with predictors.
SWAG is applied to these shuffled datasets.
The entropy of variable frequency and eigenvalue centrality is computed for the null models.
p-values are computed by comparing the SWAG network with these null models.
Small p-value (< 0.05): The selected variables are likely informative.
Large p-value (≥ 0.05): The selection may be random.
This source code is released under is the GNU AFFERO GENERAL PUBLIC LICENSE (AGPL) v3.0.
Molinari, R. et al. SWAG: A Wrapper Method for Sparse Learning (2021)