

ssdtools is an R package to fit and plot Species
Sensitivity Distributions (SSD).
SSDs are cumulative probability distributions which are fitted to toxicity concentrations for different species as described by Posthuma et al. (2001). The ssdtools package uses Maximum Likelihood to fit distributions such as the log-normal, log-logistic, log-Gumbel (also known as the inverse Weibull), gamma, Weibull and log-normal log-normal mixture. Multiple distributions can be averaged using Akaike Information Criteria. Confidence intervals on hazard concentrations and proportions are produced by bootstrapping.
ssdtools can handle censored data with two limitations.
It is currently only possible to model average when the distributions
have the same number of parameters and confidence intervals can only be
estimated using non-parametric (as opposed to parametric)
bootstrapping.
The dependency ssddata
provides example data sets for several chemicals including Boron. The ECOTOX Knowledgebase is another
source of toxicity concentrations for over 12,000 chemicals.
library(ssdtools)
ssddata::ccme_boron
#> # A tibble: 28 × 5
#> Chemical Species Conc Group Units
#> <chr> <chr> <dbl> <fct> <chr>
#> 1 Boron Oncorhynchus mykiss 2.1 Fish mg/L
#> 2 Boron Ictalurus punctatus 2.4 Fish mg/L
#> 3 Boron Micropterus salmoides 4.1 Fish mg/L
#> 4 Boron Brachydanio rerio 10 Fish mg/L
#> 5 Boron Carassius auratus 15.6 Fish mg/L
#> 6 Boron Pimephales promelas 18.3 Fish mg/L
#> 7 Boron Daphnia magna 6 Invertebrate mg/L
#> 8 Boron Opercularia bimarginata 10 Invertebrate mg/L
#> 9 Boron Ceriodaphnia dubia 13.4 Invertebrate mg/L
#> 10 Boron Entosiphon sulcatum 15 Invertebrate mg/L
#> # ℹ 18 more rowsThe six default distributions are fit using
ssd_fit_dists()
fits <- ssd_fit_dists(ssddata::ccme_boron)and can be quickly plotted using autoplot
autoplot(fits)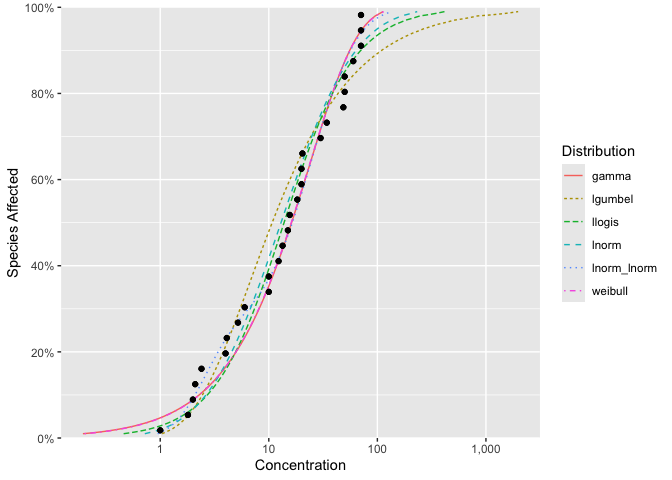
The goodness of fit can be assessed using ssd_gof
ssd_gof(fits, wt = TRUE)
#> # A tibble: 6 × 14
#> dist npars nobs log_lik aic aicc delta wt bic ad ks cvm
#> <chr> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 gamma 2 28 -117. 238. 238. 0.005 0.357 240. 0.440 0.117 0.0554
#> 2 lgumbel 2 28 -120. 244. 245. 6.56 0.013 247. 0.829 0.158 0.134
#> 3 llogis 2 28 -119. 241. 241. 3.39 0.066 244. 0.487 0.0994 0.0595
#> 4 lnorm 2 28 -118. 239. 240. 1.40 0.177 242. 0.507 0.107 0.0703
#> 5 lnorm_l… 5 28 -115. 240. 243. 4.98 0.03 247. 0.320 0.116 0.0414
#> 6 weibull 2 28 -117. 238. 238. 0 0.357 240. 0.434 0.117 0.0542
#> # ℹ 2 more variables: at_bound <lgl>, computable <lgl>and the model-averaged 5% hazard concentration estimated (with
bootstrapping to get confidence intervals) using
ssd_hc.
withr::with_seed(99, {
hc5 <- ssd_hc(fits, ci = TRUE)
})
print(hc5)
#> # A tibble: 1 × 15
#> dist proportion est se lcl ucl wt level est_method ci_method
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
#> 1 average 0.05 1.26 0.782 0.407 3.29 1 0.95 multi weighted_sa…
#> # ℹ 5 more variables: boot_method <chr>, nboot <dbl>, pboot <dbl>,
#> # dists <list>, samples <list>Model-averaged predictions complete with confidence intervals can
also be estimated by parametric bootstrapping using the
stats generic predict.
boron_pred <- predict(fits, ci = TRUE)The predictions can be plotted together with the original data using
ssd_plot.
library(ggplot2)
theme_set(theme_bw())
ssd_plot(ssddata::ccme_boron, boron_pred,
shape = "Group", color = "Group", label = "Species",
xlab = "Concentration (mg/L)", ribbon = TRUE
) +
expand_limits(x = 3000) +
scale_colour_ssd()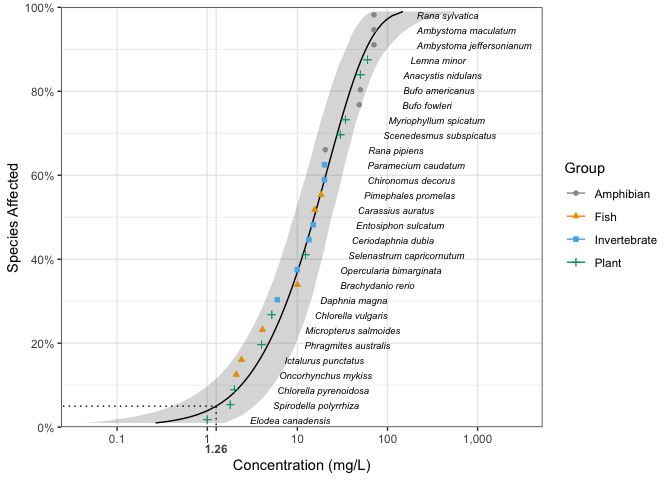
Get started with ssdtools at https://bcgov.github.io/ssdtools/articles/ssdtools.html.
A shiny app to allow non-R users to interface with ssdtools is available at https://github.com/bcgov/shinyssdtools.
For the latest changes to the development version see the NEWS file.
For recent developments in SSD modeling including a review of existing software see:
Fox, D.R., et al. 2021. Recent Developments in Species Sensitivity Distribution Modeling. Environ Toxicol Chem 40(2): 293–308. https://doi.org/10.1002/etc.4925.
To cite package 'ssdtools' in publications use:
Thorley J, Fisher R, Fox D, Schwarz C (2025). "ssdtools v2: An R
package to fit Species Sensitivity Distributions." _Journal of Open
Source Software_, *10*(105), 7492. doi:10.21105/joss.07492
<https://doi.org/10.21105/joss.07492>.
A BibTeX entry for LaTeX users is
@Article{,
title = {ssdtools v2: An R package to fit Species Sensitivity Distributions},
author = {Joe Thorley and Rebecca Fisher and David Fox and Carl Schwarz},
year = {2025},
journal = {Journal of Open Source Software},
volume = {10},
number = {105},
pages = {7492},
doi = {10.21105/joss.07492},
}To install the latest release version from CRAN.
install.packages("ssdtools")The website for the release version is at https://bcgov.github.io/ssdtools/.
To install the development version from GitHub
# install.packages("remotes")
remotes::install_github("bcgov/ssdtools")or from r-universe.
install.packages("ssdtools", repos = c("https://bcgov.r-universe.dev", "https://cloud.r-project.org"))The website for the development version is at https://bcgov.github.io/ssdtools/dev/.
To report bugs/issues/feature requests, please file an issue.
If you would like to contribute to the package, please see our CONTRIBUTING guidelines.
Please note that the ssdtools project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
Posthuma, L., Suter II, G.W., and Traas, T.P. 2001. Species Sensitivity Distributions in Ecotoxicology. CRC Press.
Copyright 2015-2023 Province of British Columbia
Copyright 2021 Environment and Climate Change Canada
Copyright 2023-2025 Australian Government Department of Climate Change,
Energy, the Environment and Water
The documentation is released under the CC BY 4.0 License
The code is released under the Apache License 2.0